python操作txt文件中数据教程[3]-python读取文件夹中所有txt文件并将数据转为csv文件
python操作txt文件中数据教程[3]-python读取文件夹中所有txt文件并将数据转为csv文件
觉得有用的话,欢迎一起讨论相互学习~
参考文献
python操作txt文件中数据教程[1]-使用python读写txt文件
python操作txt文件中数据教程[2]-python提取txt文件
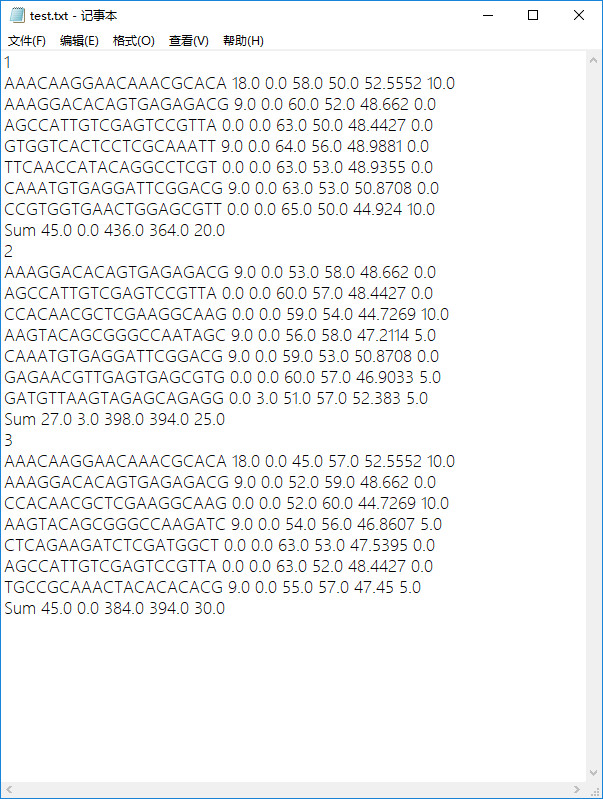
- 原始txt文件
![]()
![]()
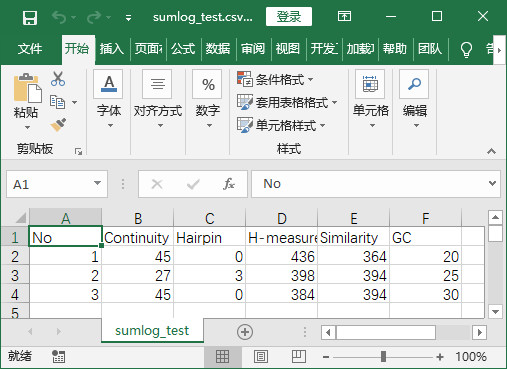
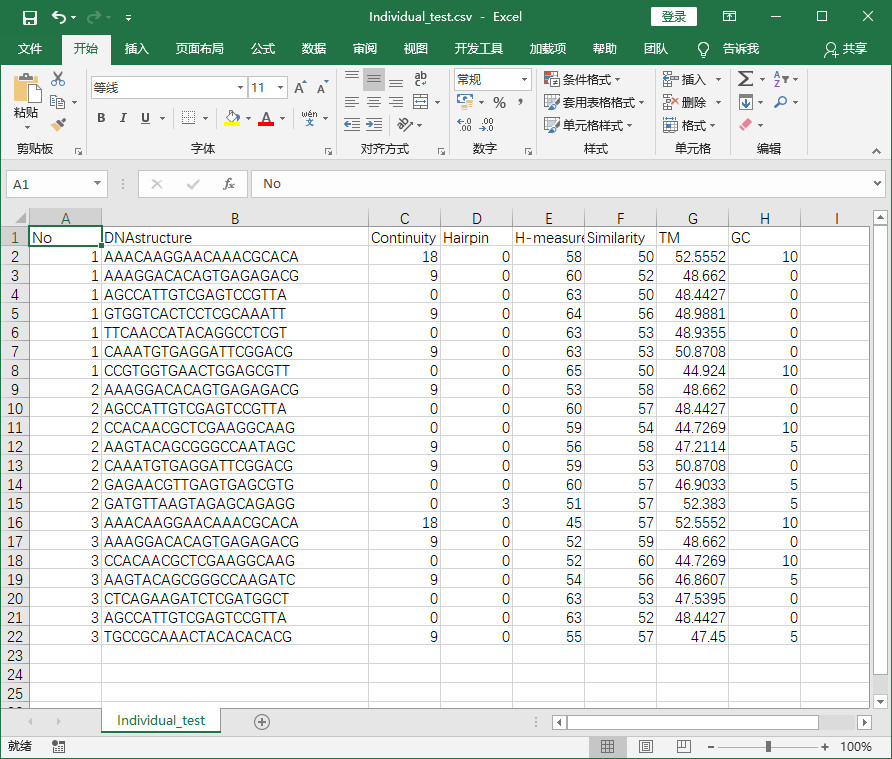
- 程序实现后结果
![]()
![]()
![]()
程序实现
import csv
import os
SUM_LOG_FILE = [] # sum_csv文件名
INDIVIDUAL_LOG_FILE = [] # individual_csv文件名
File_Name = [] # txt_文件名
DNA_Group = 7 # 表示每7条DNA组成一个组
Sum_log_file_header = ["No", "Continuity", "Hairpin", "H-measure", "Similarity", "GC"]
Individual_log_file_header = ["No", "DNAstructure", "Continuity", "Hairpin", "H-measure", "Similarity", "TM", "GC"]
def Read_Files(filename):
DNA_log = [] # 精英种群个体日志mod9=1-8
Sum_log = [] # 精英种群总体日志mod9=0
sum_evaindex = [[] for i in range(6)]
Individual_evaindex = [[] for i in range(8)]
with open(filename, 'r') as f:
i = 1
for line in f.readlines():
if i%9 == 0:
Sum_log.append(line)
else:
DNA_log.append(line)
i = i + 1
f.close()
Sum_no = 1
dna_log_no = 0
for Sum in Sum_log:
sum_eva_index = Sum.split("\n")[0].split(" ")[1:]
sum_evaindex[0].append(int(Sum_no))
sum_evaindex[1].append(float(sum_eva_index[0])) # Con
sum_evaindex[2].append(float(sum_eva_index[1])) # HP
sum_evaindex[3].append(float(sum_eva_index[2])) # Hm
sum_evaindex[4].append(float(sum_eva_index[3])) # Si
sum_evaindex[5].append(float(sum_eva_index[4])) # GC
Sum_no = Sum_no + 1
for dna_log in DNA_log:
# 获取序号值
if (dna_log_no + 1)%8 == 1:
for i in range(DNA_Group):
Individual_evaindex[0].append(int(dna_log.split("\n")[0]))
else:
# 获取各项指标
Individual_evaindex[1].append(dna_log.split("\n")[0].split(" ")[0]) # 所有DNA序列全部记载,使用原有的str字符串类型记载
Individual_evaindex[2].append(float(dna_log.split("\n")[0].split(" ")[1])) # DNA序列的连续值Con,注意要转换为浮点数类型
Individual_evaindex[3].append(float(dna_log.split("\n")[0].split(" ")[2])) # Hp茎区匹配
Individual_evaindex[4].append(float(dna_log.split("\n")[0].split(" ")[3])) # H-measure
Individual_evaindex[5].append(float(dna_log.split("\n")[0].split(" ")[4])) # Similarity
Individual_evaindex[6].append(float(dna_log.split("\n")[0].split(" ")[5])) # TM
Individual_evaindex[7].append(float(dna_log.split("\n")[0].split(" ")[6])) # GC
dna_log_no = dna_log_no + 1
return sum_evaindex, Individual_evaindex
# 将数据写入csv日志文件中
def Write_SumFiles(filename, sum_evaindex):
with open(filename, "w", newline='') as f:
writer = csv.writer(f)
writer.writerow(Sum_log_file_header) # 注意,此处使用writerow而不是使用writerows
for i in range(sum_evaindex[0][-1]):
writer.writerow(
[sum_evaindex[0][i], sum_evaindex[1][i], sum_evaindex[2][i], sum_evaindex[3][i], sum_evaindex[4][i],
sum_evaindex[5][i]])
f.close()
def Write_IndividualFiles(filename, sum_evaindex, Individual_evaindex):
with open(filename, "w", newline='') as f:
writer = csv.writer(f)
writer.writerow(Individual_log_file_header) # 注意,此处使用writerow而不是使用writerows
for i in range(sum_evaindex[0][-1]*DNA_Group):
writer.writerow(
[Individual_evaindex[0][i], Individual_evaindex[1][i], Individual_evaindex[2][i],
Individual_evaindex[3][i],
Individual_evaindex[4][i], Individual_evaindex[5][i], Individual_evaindex[6][i],
Individual_evaindex[7][i]])
f.close()
def file_name(file_dir):
for files in os.listdir(file_dir):
if os.path.splitext(files)[1] == '.txt':
File_Name.append(files)
SUM_LOG_FILE.append("./test/sumlog_" + os.path.splitext(files)[0] + ".csv")
INDIVIDUAL_LOG_FILE.append("./test/Individual_" + os.path.splitext(files)[0] + ".csv")
# 获取当前目录下所有txt文件名
file_name(".")
for i, j, k in zip(File_Name, SUM_LOG_FILE, INDIVIDUAL_LOG_FILE):
print(i)
print(j)
print(k)
Sum_Evaindex, Individual_Evaindex = Read_Files(i)
Write_SumFiles(filename=j, sum_evaindex=Sum_Evaindex)
Write_IndividualFiles(filename=k, sum_evaindex=Sum_Evaindex, Individual_evaindex=Individual_Evaindex)









 浙公网安备 33010602011771号
浙公网安备 33010602011771号