Python Machine Learning-Chapter4
Chapter4 Building Good Training Sets – Data Preprocessing
4.1 Dealing with missing data
如何判断数据框内的数据是否有空值呢?
import pandas as pd from io import StringIO csv_data = '''A, B, C, D 1.0,2.0,3.0,4.0 5.0,6.0,,8.0 10.0,11.0,12.0,''' df = pd.read_csv(StringIO(csv_data)) df.isnull() ''' A B C D 0 False False False False 1 False False True False 2 False False False True ''' df.isnull().sum() ''' A 0 B 0 C 1 D 1 dtype: int64
4.2 Eliminating samples or features with missing values
既可以去除含NA的行,也可以去除含NA的列
import pandas as pd
from io import StringIO
csv_data = '''A, B, C, D
1.0,2.0,3.0,4.0
5.0,6.0,,8.0
10.0,11.0,12.0,'''
df = pd.read_csv(StringIO(csv_data), names=['A', 'B', 'C', 'D'], header=0)
df
'''
A B C D
0 1.0 2.0 3.0 4.0
1 5.0 6.0 NaN 8.0
2 10.0 11.0 12.0 NaN
'''
df.dropna()#去除含NaN的行
'''
A B C D
0 1.0 2.0 3.0 4.0
'''
df.dropna(axis=1)#axis参数设置去除NA列
'''
A B
0 1.0 2.0
1 5.0 6.0
2 10.0 11.0
'''
# only drop rows where all columns are NaN
df.dropna(how='all')
# drop rows that have not at least 4 non-NaN values至少thresh个非NA的行才被保留
df.dropna(thresh=4)
'''
A B C D
0 1.0 2.0 3.0 4.0
'''
# only drop rows where NaN appear in specific columns (here: 'C')
df.dropna(subset=['C'])
'''
A B C D
0 1.0 2.0 3.0 4.0
2 10.0 11.0 12.0 NaN
'''
4.3 Imputing missing values
可以对NaN进行填充,用平均值或者出现次数最多的数据进行填充
import pandas as pd
from io import StringIO
csv_data = '''A,B,C,D
1.0,2.0,3.0,4.0
5.0,6.0,,8.0
10.0,11.0,12.0,'''
df = pd.read_csv(StringIO(csv_data), names=['A','B', 'C', 'D'], header=0)
from sklearn.preprocessing import Imputer
imr = Imputer(missing_values='NaN', strategy='mean', axis=0)
#strategy = 'most_frequent'用出现次数最多的数据填充NaN
#axis = 1 行的平均值填充NaN
imr = imr.fit(df)
df.values
'''
array([[ 1., 2., 3., 4.],
[ 5., 6., nan, 8.],
[10., 11., 12., nan]])
'''
imputed_data = imr.transform(df.values)
imputed_data
'''
array([[ 1. , 2. , 3. , 4. ],
[ 5. , 6. , 7.5, 8. ],
[10. , 11. , 12. , 6. ]])
'''
4.4 Handling categorical data and Mapping ordinal features
有的特征虽然不是数值型的,但是也有大小之分,比如T-shirt,XL > L > M,这样的特征称为Ordinal features,而单纯的字符串型特征称为nominal features,先创建一个新的数据框来说明这个问题:
import pandas as pd
df = pd.DataFrame([
['green', 'M', 10.1, 'class1'],
['red', 'L', 13.5, 'class2'],
['blue', 'XL', 15.3, 'class1'] ])
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
df.columns = ['color', 'size', 'price', 'classlabel']
df
'''
color size price classlabel
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
其中包含nominal feature (color), an ordinal feature (size) and a numerical feature (price),虽然ordinal feature不是数值型,但是也有大小之分,所以需要转换成数值型的特征,现在没有现成的方法完成转换,需要我们自己进行Mapping:
import pandas as pd
df = pd.DataFrame([
['green', 'M', 10.1, 'class1'],
['red', 'L', 13.5, 'class2'],
['blue', 'XL', 15.3, 'class1'] ])
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
df.columns = ['color', 'size', 'price', 'classlabel']
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
size_mapping = {'XL' :3, 'L':2, 'M':1}#XL = L +1 = M + 2
df['size'] = df['size'].map(size_mapping)
df
'''
color size price classlabel
0 green 1 10.1 class1
1 red 2 13.5 class2
2 blue 3 15.3 class1
'''
#也可以转回去,把数字型转换为字符型
str_mapping = {v:k for k,v in size_mapping.items()}
df['size'] = df['size'].map(str_mapping)
df
'''
color size price classlabel
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
许多机器学习算法库要求class label是数值型的,参考上面的mapping方法对class label进行数值转换
import pandas as pd
df = pd.DataFrame([
['green', 'M', 10.1, 'class1'],
['red', 'L', 13.5, 'class2'],
['blue', 'XL', 15.3, 'class1'] ])
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
df.columns = ['color', 'size', 'price', 'classlabel']
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
size_mapping = {'XL' :3, 'L':2, 'M':1}#XL = L +1 = M + 2
df['size'] = df['size'].map(size_mapping)
df
'''
color size price classlabel
0 green 1 10.1 class1
1 red 2 13.5 class2
2 blue 3 15.3 class1
'''
#也可以转回去,把数字型转换为字符型
str_mapping = {v:k for k,v in size_mapping.items()}
df['size'] = df['size'].map(str_mapping)
df
'''
color size price classlabel
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
import numpy as np
class_mapping = {label:idx for idx, label in enumerate(np.unique(df['classlabel']))}
class_mapping
'''
{'class1': 0, 'class2': 1}
'''
df['classlabel'] = df['classlabel'].map(class_mapping)
df
'''
color size price classlabel
0 green M 10.1 0
1 red L 13.5 1
2 blue XL 15.3 0
'''
#也可以转回去,将数字型label转换为字符型
inv_class_mapping = {v: k for k, v in class_mapping.items()}
df['classlabel'] = df['classlabel'].map(inv_class_mapping)
df
'''
color size price classlabel
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
当然sklearn有内置的方法(LabelEncoder class)进行label数字型和字符型互相转换:
import pandas as pd
df = pd.DataFrame([
['green', 'M', 10.1, 'class1'],
['red', 'L', 13.5, 'class2'],
['blue', 'XL', 15.3, 'class1'] ])
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
df.columns = ['color', 'size', 'price', 'classlabel']
df
'''
0 1 2 3
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
size_mapping = {'XL' :3, 'L':2, 'M':1}#XL = L +1 = M + 2
df['size'] = df['size'].map(size_mapping)
df
'''
color size price classlabel
0 green 1 10.1 class1
1 red 2 13.5 class2
2 blue 3 15.3 class1
'''
#也可以转回去,把数字型转换为字符型
str_mapping = {v:k for k,v in size_mapping.items()}
df['size'] = df['size'].map(str_mapping)
df
'''
color size price classlabel
0 green M 10.1 class1
1 red L 13.5 class2
2 blue XL 15.3 class1
'''
from sklearn.preprocessing import LabelEncoder
class_le = LabelEncoder()
y = class_le.fit_transform(df['classlabel'].values)
y
'''
array([0, 1, 0], dtype=int64)
'''
class_le.inverse_transform(y)
'''
array(['class1', 'class2', 'class1'], dtype=object)
'''
4.5 Performing one-hot encoding on nominal features
nominal feature同样需要进行encode转换成数值,但是如果采用和ordinal feature同样的处理方式:
#coding=utf-8
import pandas as pd
df = pd.DataFrame([
['green', 'M', 10.1, 'class1'],
['red', 'L', 13.5, 'class2'],
['blue', 'XL', 15.3, 'class1'] ])
df.columns = ['color', 'size', 'price', 'classlabel']
size_mapping = {'XL' :3, 'L':2, 'M':1}#XL = L +1 = M + 2
df['size'] = df['size'].map(size_mapping)
df
'''
color size price classlabel
0 green 1 10.1 class1
1 red 2 13.5 class2
2 blue 3 15.3 class1
'''
from sklearn.preprocessing import LabelEncoder
class_le = LabelEncoder()
y = class_le.fit_transform(df['classlabel'].values)
y#array([0, 1, 0], dtype=int64)
'''
#也可以转回去
class_le.inverse_transform(y)
array(['class1', 'class2', 'class1'], dtype=object)
'''
X = df[['color', 'size', 'price']].values
color_le = LabelEncoder()
X[:, 0] = color_le.fit_transform(X[:, 0])
X
'''
array([[1, 1, 10.1],
[2, 2, 13.5],
[0, 3, 15.3]], dtype=object)
blue -> 0
green -> 1
red -> 2
'''
如果把这样的数据给模型训练,模型会认为green特征大于blue,red特征大于green特征,这明显是错误的,所以nominal特征不能采用和ordinal特征一样的处理方式。处理nominal features常用的方法是one-hot encoding,如将color特征变成多维特征,如a blue sample can be encoded as blue=1, green=0, red=0:OneHot Encoder 内置于sklearn.preprocessing中:
#coding=utf-8
import pandas as pd
df = pd.DataFrame([
['green', 'M', 10.1, 'class1'],
['red', 'L', 13.5, 'class2'],
['blue', 'XL', 15.3, 'class1'] ])
df.columns = ['color', 'size', 'price', 'classlabel']
size_mapping = {'XL' :3, 'L':2, 'M':1}#XL = L +1 = M + 2
df['size'] = df['size'].map(size_mapping)
df
'''
color size price classlabel
0 green 1 10.1 class1
1 red 2 13.5 class2
2 blue 3 15.3 class1
'''
from sklearn.preprocessing import LabelEncoder
class_le = LabelEncoder()
y = class_le.fit_transform(df['classlabel'].values)
y#array([0, 1, 0], dtype=int64)
'''
#也可以转回去
class_le.inverse_transform(y)
array(['class1', 'class2', 'class1'], dtype=object)
'''
X = df[['color', 'size', 'price']].values
color_le = LabelEncoder()
X[:, 0] = color_le.fit_transform(X[:, 0])
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder(categorical_features=[0])
print(ohe.fit_transform(X).toarray())
'''
array([[ 0. , 1. , 0. , 1. , 10.1],
[ 0. , 0. , 1. , 2. , 13.5],
[ 1. , 0. , 0. , 3. , 15.3]])
'''
#更常用的one-hot encoding方法是利用pandas中的get_dummies方法,get_dummies方法会只对string columns进行ond-hot encoding,而其他columns不会更改
pd.get_dummies(df[['price', 'color', 'size']])
'''
price size color_blue color_green color_red
0 10.1 1 0 1 0
1 13.5 2 0 0 1
2 15.3 3 1 0 0
'''
categorical_features参数指定变量的列(想要进行one-hot encoding的特征位置),而transform方法返回一个稀疏矩阵。
4.6 Partitioning a dataset in training and test sets
#coding=utf-8
import pandas as pd
import numpy as np
df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data', header=None)
df_wine.columns =[
'Class label', 'Alcohol',
'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium',
'Total phenols', 'Flavanoids',
'Nonflavanoid phenols',
'Proanthocyanins',
'Color intensity', 'Hue',
'OD280/OD315 of diluted wines',
'Proline'
]
print("Class labels", np.unique(df_wine['Class label']))
print(df_wine.head())
from sklearn.cross_validation import train_test_split
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values#通过行号,列号获取数据
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
print(X,y)
4.7 Bringing features onto the same scale
数据标准化:normalization和standardization
normalization:\( x_{norm} = (x-x_{min})/(x_{max}-x_{min}) \),sklearn.preprocessing中的MinMaxScaler方法可以进行normalization
#coding=utf-8
import pandas as pd
import numpy as np
df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data', header=None)
df_wine.columns =[
'Class label', 'Alcohol',
'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium',
'Total phenols', 'Flavanoids',
'Nonflavanoid phenols',
'Proanthocyanins',
'Color intensity', 'Hue',
'OD280/OD315 of diluted wines',
'Proline'
]
print("Class labels", np.unique(df_wine['Class label']))
print(df_wine.head())
from sklearn.cross_validation import train_test_split
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values#通过行号,列号获取数据
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
from sklearn.preprocessing import MinMaxScaler
mms = MinMaxScaler()
X_train_norm = mms.fit_transform(X_train)
X_test_norm = mms.transform(X_test)
print(X_train_norm, X_test_norm)
standardization:\( x_{std} = (x - \mu_{x})/\sigma_x \),其中\( \mu_{x}\)是某特征的mean,\( \sigma_x\)是某特征的standard deviation:
#coding=utf-8
import pandas as pd
import numpy as np
df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data', header=None)
df_wine.columns =[
'Class label', 'Alcohol',
'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium',
'Total phenols', 'Flavanoids',
'Nonflavanoid phenols',
'Proanthocyanins',
'Color intensity', 'Hue',
'OD280/OD315 of diluted wines',
'Proline'
]
print("Class labels", np.unique(df_wine['Class label']))
print(df_wine.head())
from sklearn.cross_validation import train_test_split
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values#通过行号,列号获取数据
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
print("============MinMaxScaler============")
from sklearn.preprocessing import MinMaxScaler
mms = MinMaxScaler()
X_train_norm = mms.fit_transform(X_train)
X_test_norm = mms.transform(X_test)
print(X_train_norm, X_test_norm)
print("============StandardScaler============")
from sklearn.preprocessing import StandardScaler
stdsc = StandardScaler()
X_train_std = stdsc.fit_transform(X_train)
X_test_std = stdsc.transform(X_test)
print(X_train_std,X_test_std)
norm和std方法的差别如下表(考虑输入是0到5):
4.8 Selecting meaningful features
过拟合的解决方法:
- Collect more training data(不现实,训练数据有限)
- Introduce a penalty for complexity via regularization
- Choose a simpler model with fewer parameters
- Reduce the dimensionality of the data
4.9 Sequential feature selection algorithms(时序特征)
主要有两种维度减少的方法:特征选择和特征提取,下面以特征选择为例
#coding=utf-8
import pandas as pd
import numpy as np
from sklearn.base import clone
from itertools import combinations
import numpy as np
from sklearn.cross_validation import train_test_split
from sklearn.metrics import accuracy_score
class SBS():
def __init__(self, estimator, k_features, scoring=accuracy_score, test_size=0.25, random_state=1):
self.scoring = scoring
self.estimator = clone(estimator)
self.k_features = k_features
self.test_size = test_size
self.random_state = random_state
def fit(self, X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=self.test_size, random_state=self.random_state)
dim = X_train.shape[1]
self.indices_ = tuple(range(dim))
self.subsets_ = [self.indices_]
score = self._calc_score(X_train, y_train, X_test, y_test, self.indices_)
self.scores_ = [score]
while dim > self.k_features:
scores = []
subsets = []
for p in combinations (self.indices_, r=dim-1):
score = self._calc_score(X_train, y_train, X_test, y_test, p)
scores.append(score)
subsets.append(p)
best = np.argmax(scores)
self.indices_ = subsets[best]
self.subsets_.append(self.indices_)
dim -= 1
self.scores_.append(scores[best])
self.k_score_ = self.scores_[-1]
return self
def transform(self, X):
return X[:, self.indices_]
def _calc_score(self, X_train, y_train, X_test, y_test, indices):
self.estimator.fit(X_train[:, indices], y_train)
y_pred = self.estimator.predict(X_test[:, indices])
score = self.scoring(y_test, y_pred)
return score
df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data', header=None)
df_wine.columns =[
'Class label', 'Alcohol',
'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium',
'Total phenols', 'Flavanoids',
'Nonflavanoid phenols',
'Proanthocyanins',
'Color intensity', 'Hue',
'OD280/OD315 of diluted wines',
'Proline'
]
print("Class labels", np.unique(df_wine['Class label']))
print(df_wine.head())
from sklearn.cross_validation import train_test_split
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values#通过行号,列号获取数据
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
print("============MinMaxScaler============")
from sklearn.preprocessing import MinMaxScaler
mms = MinMaxScaler()
X_train_norm = mms.fit_transform(X_train)
X_test_norm = mms.transform(X_test)
print(X_train_norm, X_test_norm)
print("============StandardScaler============")
from sklearn.preprocessing import StandardScaler
stdsc = StandardScaler()
X_train_std = stdsc.fit_transform(X_train)
X_test_std = stdsc.transform(X_test)
print(X_train_std,X_test_std)
from sklearn.neighbors import KNeighborsClassifier
import matplotlib.pyplot as plt
knn = KNeighborsClassifier(n_neighbors=2)
sbs = SBS(knn, k_features=1)
sbs.fit(X_train_std, y_train)
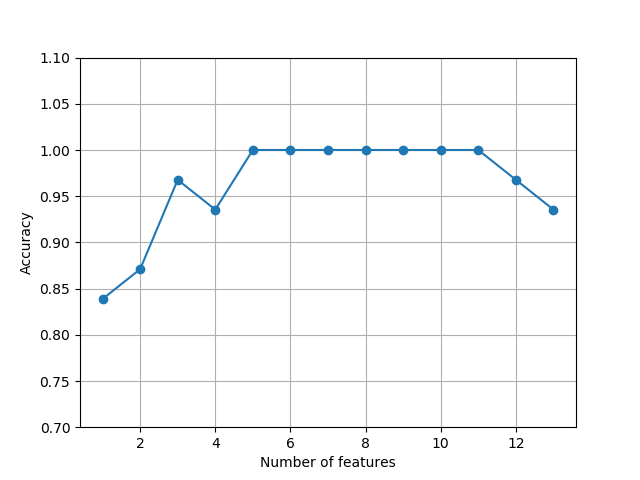
#plot the classification accuracy of the KNN classifier that was calculated on the validation dataset.
k_feat = [len(k) for k in sbs.subsets_]
plt.plot(k_feat, sbs.scores_, marker='o')
plt.ylim([0.7, 1.1])
plt.ylabel('Accuracy')
plt.xlabel('Number of features')
plt.grid()
plt.show()
#what those five features are that yielded such a good performance on the validation dataset
k5 = list(sbs.subsets_[8])
print(df_wine.columns[1:][k5])#Index(['Alcohol', 'Malic acid', 'Alcalinity of ash', 'Hue', 'Proline'], dtype='object')
#let's evaluate the performance of the KNN classifier on the original test set
knn.fit(X_train_std, y_train)
print("Training accuracy:", knn.score(X_train_std, y_train))
#Training accuracy: 0.9838709677419355
print("Test accuracy:", knn.score(X_test_std, y_test))
#Test accuracy: 0.9444444444444444
'''
We used the complete feature set and obtained ~98.4 percent accuracy on the training dataset.
However, the accuracy on the test dataset was slightly lower (~94.4 percent),
which is an indicator of a slight degree of overfitting.
'''
#Now let's use the selected 5-feature subset and see how well KNN performs
knn.fit(X_train_std[:, k5], y_train)
print('Training accuracy:', knn.score(X_train_std[:, k5], y_train))
#Training accuracy: 0.9596774193548387
print("Test accuracy:", knn.score(X_test_std[:, k5], y_test))
#Test accuracy: 0.9629629629629629
'''
Using fewer than half of the original features in the Wine dataset, the prediction
accuracy on the test set improved by almost 2 percent. Also, we reduced overfitting,
which we can tell from the small gap between test (~96.3 percent) and training
(~96.0 percent) accuracy.
'''
itertools中的combinations可以帮助我们从一个元组中选择若干个元素,并将其展示出来,如\C_6^2\)
S = ('ABCDEF')
for each in combinations(S, r=2):
print(each)
#r指定选几个
'''
('A', 'B')
('A', 'C')
('A', 'D')
('A', 'E')
('A', 'F')
('B', 'C')
('B', 'D')
('B', 'E')
('B', 'F')
('C', 'D')
('C', 'E')
('C', 'F')
('D', 'E')
('D', 'F')
('E', 'F')
'''
关于itertools包的使用,可以参考:https://docs.python.org/2/library/itertools.html
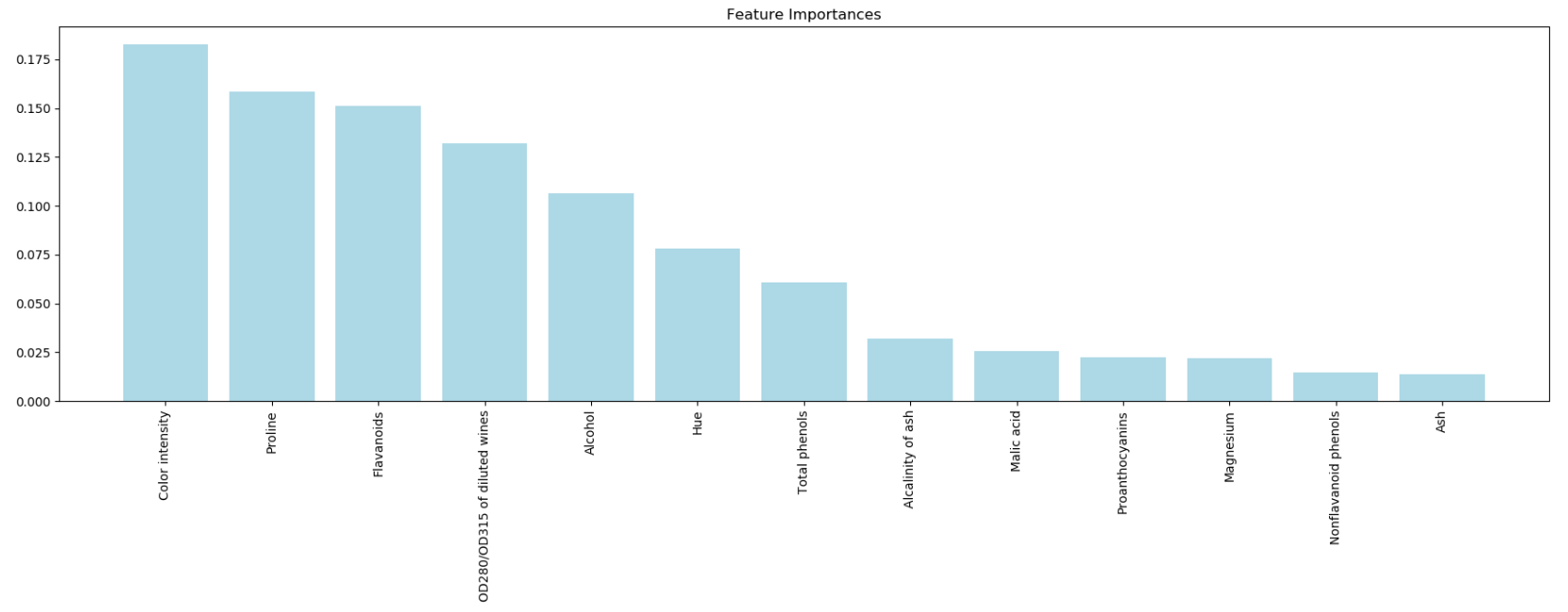
4.10 Assessing feature importance with random forests
Train a forest of 10,000 trees on the Wine dataset and rank the 13 features by their respective importance measures and we don't need to use standardized or normalized tree-based models.
#coding=utf-8
import pandas as pd
import numpy as np
df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data', header=None)
df_wine.columns =[
'Class label', 'Alcohol',
'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium',
'Total phenols', 'Flavanoids',
'Nonflavanoid phenols',
'Proanthocyanins',
'Color intensity', 'Hue',
'OD280/OD315 of diluted wines',
'Proline'
]
print("Class labels", np.unique(df_wine['Class label']))
print(df_wine.head())
from sklearn.cross_validation import train_test_split
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values#通过行号,列号获取数据
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
from sklearn.ensemble import RandomForestClassifier
feat_labels = df_wine.columns[1:]
forest = RandomForestClassifier(n_estimators=10000, random_state=0, n_jobs=-1)
forest.fit(X_train, y_train)
importances = forest.feature_importances_
indices = np.argsort(importances)[: : -1]
for f in range(X_train.shape[1]):
print("%2d) %-*s %f " % (f + 1, 60, feat_labels[indices[f]], importances[indices[f]]))
import matplotlib.pyplot as plt
plt.title('Feature Importances')
plt.bar(range(X_train.shape[1]), importances[indices], color='lightblue', align='center')
plt.xticks(range(X_train.shape[1]), feat_labels[indices], rotation=90)
plt.xlim([-1, X_train.shape[1]])
plt.tight_layout()
plt.show()
'''
we created a plot that ranks the different features in the Wine dataset
by their relative importance; note that the feature importances are normalized so that they sum up to 1.0.
'''


 浙公网安备 33010602011771号
浙公网安备 33010602011771号