R语言学习笔记(十六):处理缺失值
#识别缺失值
install.packages("VIM")
data(sleep,package="VIM")
#列出没有缺失值的行
sleep[complete.cases(sleep),]
#列出有一个或多个缺失值的行
sleep[!complete.cases(sleep),]
#有多少个缺失值
sum(is.na(sleep$Dream))
#sleep$dream上有百分之几的数据是有缺失值的
mean(is.na(sleep$Dream))
#数据集中多个行包含缺失值
mean(!complete.cases(sleep))
#探索缺失值
install.packages("mice")
library(mice)
data(sleep,package="VIM")
md.pattern(sleep)
#图形探索
library("VIM")
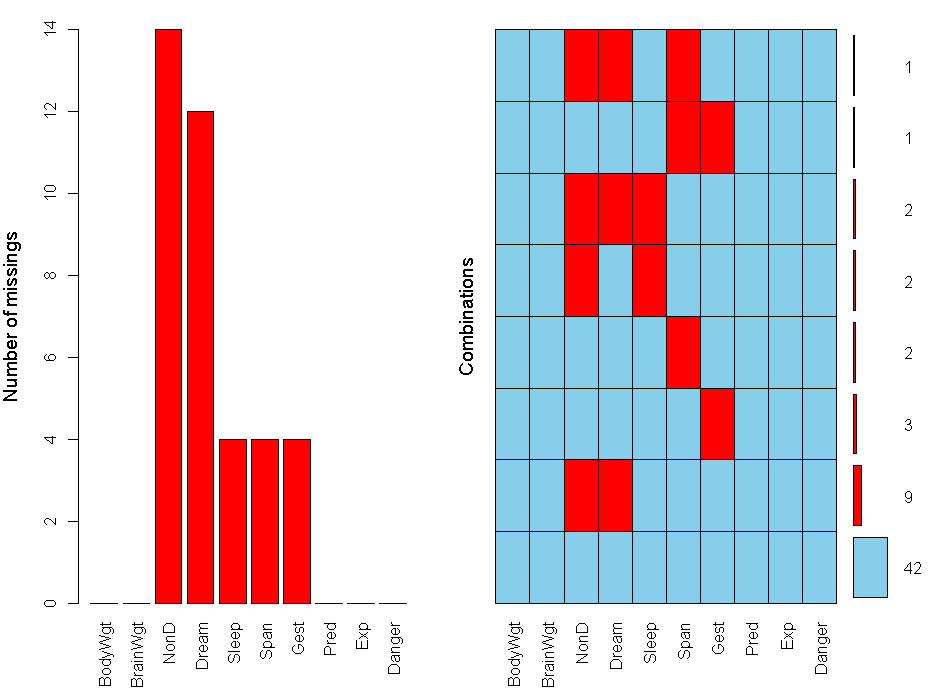
aggr(sleep,prop=FALSE,numbers=TRUE)
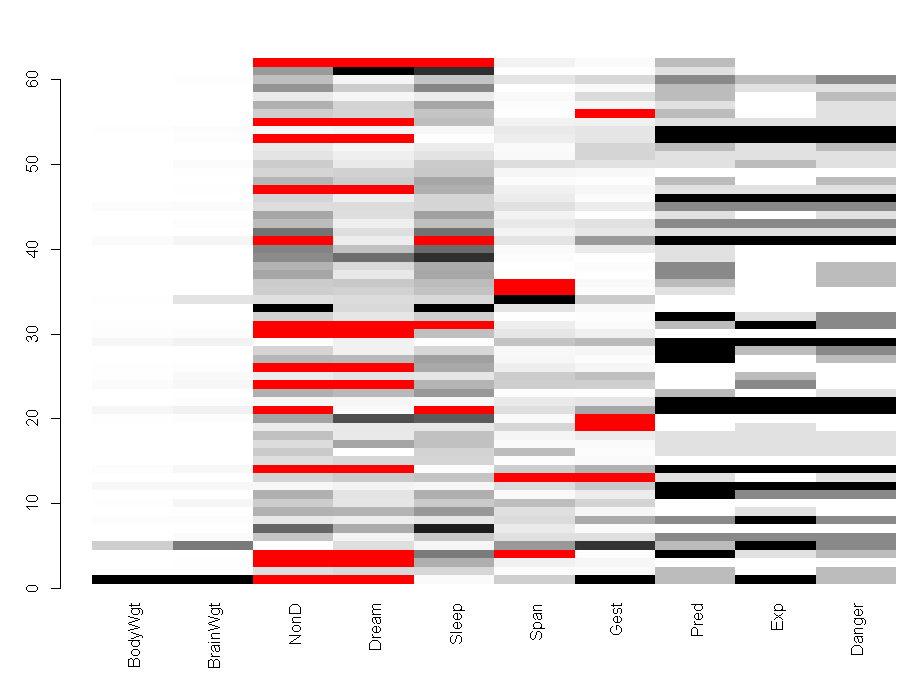
matrixplot(sleep)
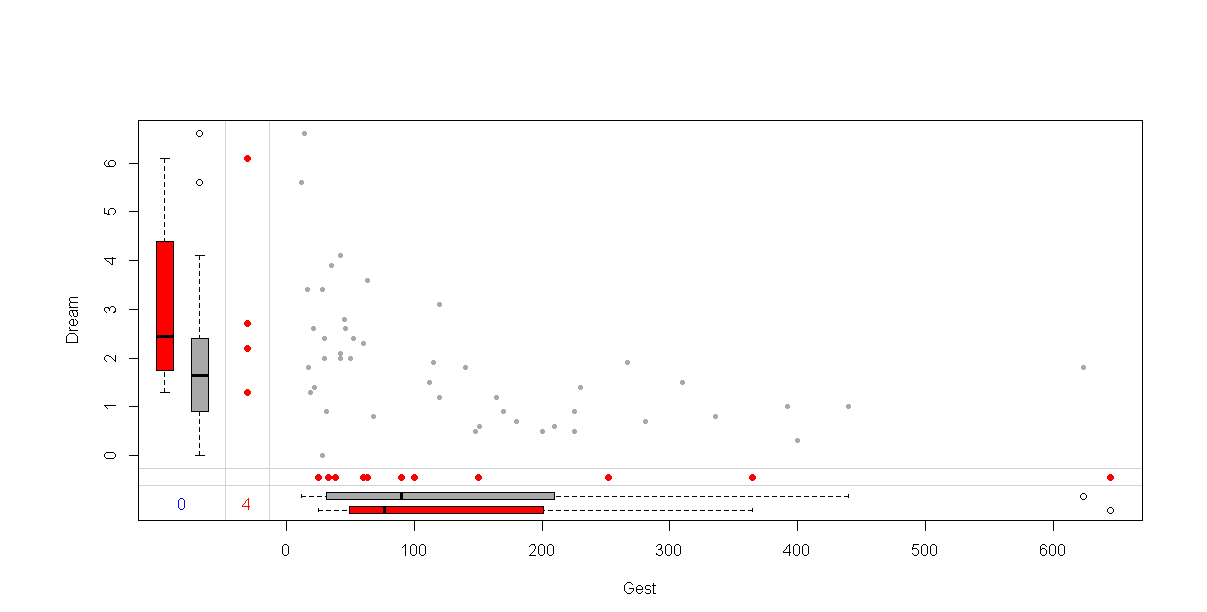
marginplot(sleep[c("Gest","Dream")],pch=c(20),col=c("darkgray","red","blue"))
#用相关性探索缺失值 x<-as.data.frame(abs(is.na(sleep))) head(sleep,n=5) head(x,n=5) y<-x[which(apply(x,2,sum)>0)] cor(y)
NonD Dream Sleep Span Gest
NonD 1.00 0.91 0.49 0.02 -0.14
Dream 0.91 1.00 0.20 0.04 -0.13
Sleep 0.49 0.20 1.00 -0.07 -0.07
Span 0.02 0.04 -0.07 1.00 0.20
Gest -0.14 -0.13 -0.07 0.20 1.00
cor(sleep,y,use="pairwise.complete.obs")
NonD Dream Sleep Span Gest
BodyWgt 0.23 0.22 0.002 -0.06 -0.05
BrainWgt 0.18 0.16 0.008 -0.08 -0.07
NonD NA NA NA -0.04 -0.05
Dream -0.19 NA -0.189 0.12 0.23
Sleep -0.08 -0.08 NA 0.10 0.04
Span 0.08 0.06 0.005 NA -0.07
Gest 0.20 0.05 0.160 -0.17 NA
Pred 0.05 -0.07 0.202 0.02 -0.20
Exp 0.25 0.13 0.261 -0.19 -0.19
Danger 0.07 -0.07 0.209 -0.07 -0.20
#行删除 newdata<-mydata[complete.cases(mydata),] newdata<-na.omit(mydata) options(digits=1) cor(na.omit(sleep)) fit<-lm(Dream~Span+Gest,data=na.omit(sleep)) summary(fit) #多重插补 library(mice) data(sleep,package="VIM") imp<-mice(sleep,seed=1234) fit<-with(imp,lm(Dream~Span+Gest)) pooled<-pool(fit) summary(pooled) imp imp$imp$Dream dataset3<-complete(imp,action=3) dataset3 #多重删补后的结果
BodyWgt BrainWgt NonD Dream Sleep Span Gest Pred Exp Danger
1 7e+03 6e+03 2 0.5 3 39 645 3 5 3
2 1e+00 7e+00 6 2.0 8 4 42 3 1 3
3 3e+00 4e+01 11 1.5 12 14 60 1 1 1
4 9e-01 6e+00 13 3.4 16 2 25 5 2 3
5 3e+03 5e+03 2 1.8 4 69 624 3 5 4
6 1e+01 2e+02 9 0.7 10 27 180 4 4 4
7 2e-02 3e-01 16 3.9 20 19 35 1 1 1
8 2e+02 2e+02 5 1.0 6 30 392 4 5 4
9 3e+00 3e+01 11 3.6 14 28 63 1 2 1
10 5e+01 4e+02 8 1.4 10 50 230 1 1 1
11 4e-01 6e+00 11 1.5 12 7 112 5 4 4
12 5e+02 4e+02 3 0.7 4 30 281 5 5 5
13 6e-01 2e+00 8 2.7 10 18 46 2 1 2
14 2e+02 4e+02 3 0.5 3 40 365 5 5 5
15 7e-02 1e+00 6 2.1 8 4 42 1 1 1
16 3e+00 2e+01 9 0.0 9 50 28 2 2 2
17 8e-01 4e+00 7 4.1 11 6 42 2 2 2
18 2e-01 5e+00 10 1.2 11 10 120 2 2 2
19 1e+00 2e+01 5 1.3 6 34 28 1 2 1
20 6e+01 8e+01 12 6.1 18 7 21 1 1 1
21 5e+02 7e+02 11 0.3 11 28 400 5 5 5
22 3e+01 1e+02 3 0.5 4 20 148 5 5 5
23 1e-01 1e+00 11 3.4 14 4 16 3 1 2
24 2e+02 4e+02 8 3.6 12 39 252 1 4 1
25 8e+01 3e+02 5 1.5 6 41 310 1 3 1
26 4e+01 1e+02 11 2.0 13 16 63 1 1 1
27 1e-01 4e+00 10 3.4 14 9 28 5 1 3
28 1e+00 6e+00 7 0.8 8 8 68 5 3 4
29 5e+02 7e+02 2 0.8 3 46 336 5 5 5
30 1e+02 2e+02 7 3.4 11 22 100 1 1 1
31 4e+01 6e+01 3 0.6 4 16 33 3 5 4
32 5e-03 1e-01 8 1.4 9 3 22 5 2 4
33 1e-02 2e-01 18 2.0 20 24 50 1 1 1
34 6e+01 1e+03 6 1.9 8 100 267 1 1 1
35 1e-01 3e+00 8 2.4 11 13 30 2 1 1
36 1e+00 8e+00 8 2.8 11 4 45 3 1 3
37 2e-02 4e-01 12 1.3 13 3 19 4 1 3
38 5e-02 3e-01 11 2.0 13 2 30 4 1 3
39 2e+00 6e+00 14 5.6 19 5 12 2 1 1
40 4e+00 1e+01 14 3.1 17 6 120 2 1 1
41 2e+02 5e+02 8 1.0 8 24 440 5 5 5
42 5e-01 2e+01 15 1.8 17 12 140 2 2 2
43 1e+01 1e+02 10 0.9 11 20 170 4 4 4
44 2e+00 1e+01 12 1.8 14 13 17 2 1 2
45 2e+02 2e+02 6 1.9 8 27 115 4 4 4
46 2e+00 1e+01 8 0.9 8 18 31 5 5 5
47 4e+00 4e+01 11 1.5 12 14 63 2 2 2
48 3e-01 2e+00 11 2.6 13 5 21 3 1 3
49 4e+00 5e+01 7 2.4 10 10 52 1 1 1
50 7e+00 2e+02 8 1.2 10 29 164 2 3 2
51 8e-01 1e+01 6 0.9 7 7 225 2 2 2
52 4e+00 2e+01 5 0.5 5 6 225 3 2 3
53 1e+01 1e+02 2 0.5 3 17 150 5 5 5
54 6e+01 2e+02 3 0.6 4 20 151 5 5 5
55 1e+00 1e+01 8 2.6 11 13 90 2 2 2
56 6e-02 1e+00 8 2.2 10 4 100 3 1 2
57 9e-01 3e+00 11 2.3 13 4 60 2 1 2
58 2e+00 1e+01 5 0.5 5 8 200 3 1 3
59 1e-01 2e+00 13 2.6 16 2 46 3 2 2
60 4e+00 6e+01 10 0.6 10 24 210 4 3 4
61 4e+00 4e+00 13 6.6 19 3 14 2 1 1
62 4e+00 2e+01 18 0.5 19 13 38 3 1 1
#成对删除 cor(sleep,use="pairwise.complete.obs")





