20240117进度汇报
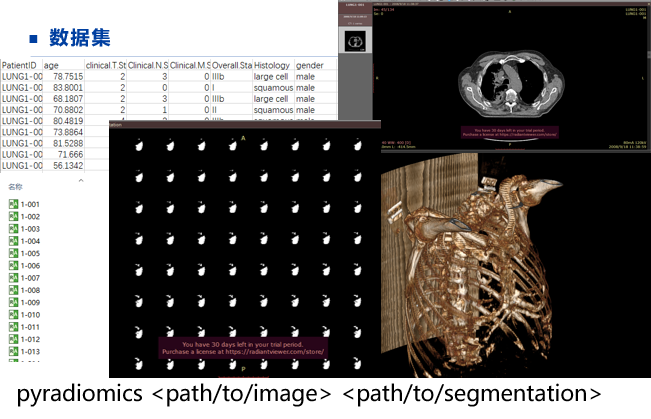
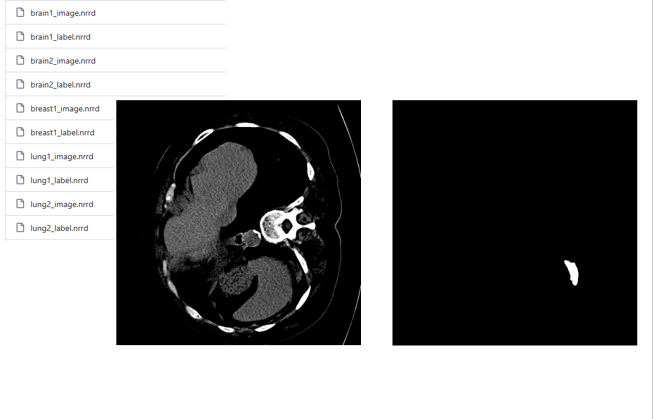

#!/usr/bin/env python from __future__ import print_function import collections import csv import logging import os import SimpleITK as sitk import radiomics from radiomics import featureextractor def main(): outPath = r'' inputCSV = os.path.join(outPath, 'testCases.csv') outputFilepath = os.path.join(outPath, 'radiomics_features.csv') progress_filename = os.path.join(outPath, 'pyrad_log.txt') params = os.path.join(outPath, 'exampleSettings', 'Params.yaml') # Configure logging rLogger = logging.getLogger('radiomics') # Set logging level # rLogger.setLevel(logging.INFO) # Not needed, default log level of logger is INFO # Create handler for writing to log file handler = logging.FileHandler(filename=progress_filename, mode='w') handler.setFormatter(logging.Formatter('%(levelname)s:%(name)s: %(message)s')) rLogger.addHandler(handler) # Initialize logging for batch log messages logger = rLogger.getChild('batch') # Set verbosity level for output to stderr (default level = WARNING) radiomics.setVerbosity(logging.INFO) logger.info('pyradiomics version: %s', radiomics.__version__) logger.info('Loading CSV') flists = [] try: with open(inputCSV, 'r') as inFile: cr = csv.DictReader(inFile, lineterminator='\n') flists = [row for row in cr] except Exception: logger.error('CSV READ FAILED', exc_info=True) logger.info('Loading Done') logger.info('Patients: %d', len(flists)) if os.path.isfile(params): extractor = featureextractor.RadiomicsFeatureExtractor(params) else: # Parameter file not found, use hardcoded settings instead settings = {} settings['binWidth'] = 25 settings['resampledPixelSpacing'] = None # [3,3,3] settings['interpolator'] = sitk.sitkBSpline settings['enableCExtensions'] = True extractor = featureextractor.RadiomicsFeatureExtractor(**settings) extractor.enableInputImages(wavelet= {'level': 2}) logger.info('Enabled input images types: %s', extractor.enabledImagetypes) logger.info('Enabled features: %s', extractor.enabledFeatures) logger.info('Current settings: %s', extractor.settings) headers = None for idx, entry in enumerate(flists, start=1): logger.info("(%d/%d) Processing Patient (Image: %s, Mask: %s)", idx, len(flists), entry['Image'], entry['Mask']) imageFilepath = entry['Image'] maskFilepath = entry['Mask'] label = entry.get('Label', None) if str(label).isdigit(): label = int(label) else: label = None if (imageFilepath is not None) and (maskFilepath is not None): featureVector = collections.OrderedDict(entry) featureVector['Image'] = os.path.basename(imageFilepath) featureVector['Mask'] = os.path.basename(maskFilepath) try: featureVector.update(extractor.execute(imageFilepath, maskFilepath, label)) with open(outputFilepath, 'a') as outputFile: writer = csv.writer(outputFile, lineterminator='\n') if headers is None: headers = list(featureVector.keys()) writer.writerow(headers) row = [] for h in headers: row.append(featureVector.get(h, "N/A")) writer.writerow(row) except Exception: logger.error('FEATURE EXTRACTION FAILED', exc_info=True) if __name__ == '__main__': main()
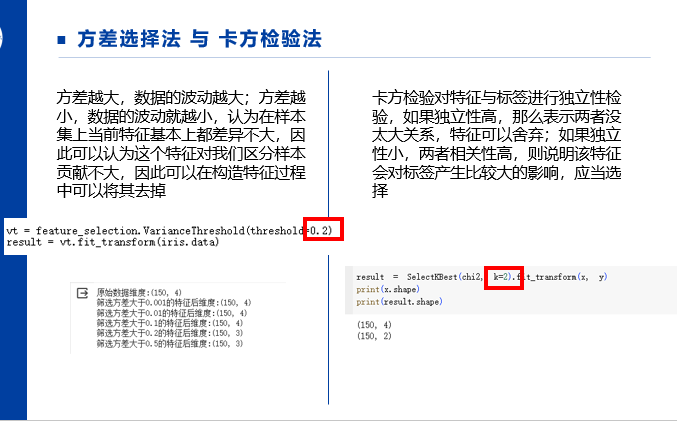
from sklearn import feature_selection from sklearn import datasets # 初始化鸢尾花数据集 iris = datasets.load_iris() # 初始化转换器(指定方差为0.2) vt = feature_selection.VarianceThreshold(threshold=0.2) # 使用转换器对数据进行低方差过滤 result = vt.fit_transform(iris.data) # 打印数据特征 print(result) print(result.shape)
from sklearn.datasets import load_iris from sklearn.feature_selection import SelectKBest from sklearn.feature_selection import chi2 #load data iris = load_iris() Y = iris.target X = iris.data print(X.shape) X_new = SelectKBest(chi2, k=2).fit_transform(X, Y) print(X_new.shape)
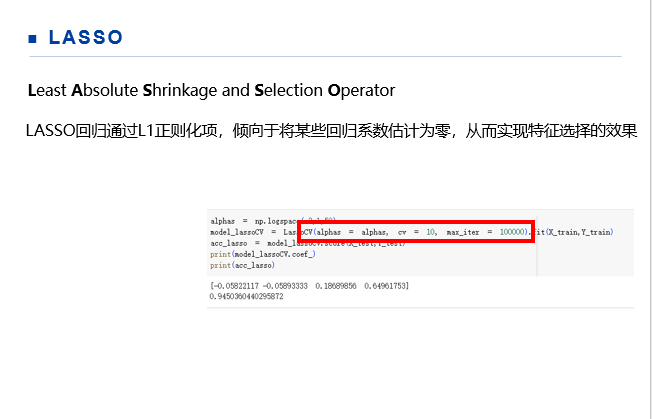
import sklearn from sklearn.linear_model import Lasso,LassoCV,LassoLarsCV from sklearn.datasets import load_iris from sklearn.model_selection import train_test_split import pandas as pd import numpy as np iris = load_iris() Y = iris.target X = iris.data X_train,X_test,Y_train,Y_test = train_test_split(X,Y,test_size=0.3) alphas = np.logspace(-3,1,50) model_lassoCV = LassoCV(alphas = alphas, cv = 10, max_iter = 100000).fit(X_train,Y_train) acc_lasso = model_lassoCV.score(X_test,Y_test) print(model_lassoCV.coef_) print(acc_lasso)
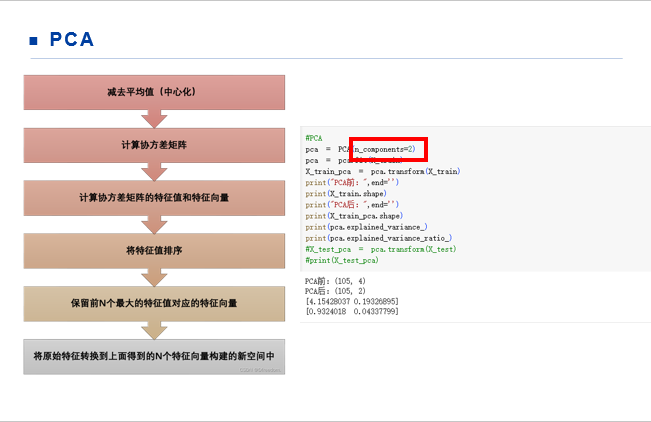
import matplotlib.pyplot as plt from sklearn.datasets import load_iris from sklearn.decomposition import PCA from sklearn.model_selection import train_test_split from sklearn import svm import pandas as pd #load data iris = load_iris() Y = iris.target print(Y) X = iris.data print(X) X_train,X_test,Y_train,Y_test = train_test_split(X,Y,test_size=0.3) #PCA pca = PCA(n_components=0.99) pca = pca.fit(X_train) print(pca.explained_variance_) print(pca.explained_variance_ratio_) X_train_pca = pca.transform(X_train) print(X_train_pca) X_test_pca = pca.transform(X_test) print(X_test_pca) #SVM lin_svc = svm.SVC(kernel='linear').fit(X_train_pca,Y_train) rbf_svc = svm.SVC(kernel='rbf').fit(X_train_pca,Y_train) poly_svc = svm.SVC(kernel='poly',degree=3).fit(X_train_pca,Y_train) lin_svc_pre = lin_svc.predict(X_test_pca) rbf_svc_pre = rbf_svc.predict(X_test_pca) poly_svc_pre = poly_svc.predict(X_test_pca) acc_lin_svc = lin_svc.score(X_test_pca,Y_test) acc_rbf_svc = rbf_svc.score(X_test_pca,Y_test) acc_poly_svc = poly_svc.score(X_test_pca,Y_test) print('acc_lin_svc: ',acc_lin_svc) print('acc_lin_predicted: ',lin_svc_pre) print('acc_rbf_svc: ',acc_rbf_svc) print('acc_rbf_predicted: ',rbf_svc_pre) print('acc_poly_svc: ',acc_poly_svc) print('acc_poly_predicted: ',poly_svc_pre)

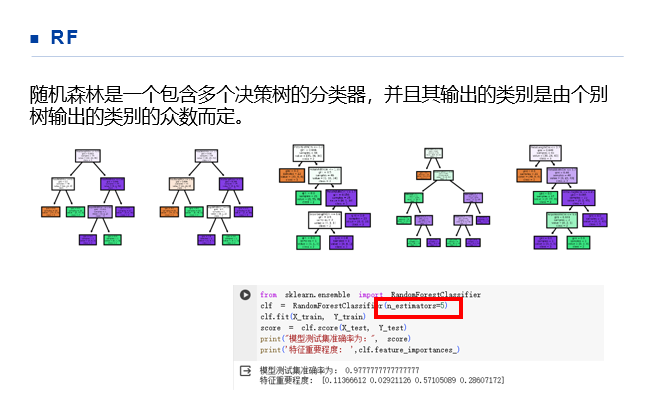
import matplotlib.pyplot as plt from sklearn.datasets import load_iris from sklearn.decomposition import PCA from sklearn.model_selection import train_test_split from sklearn import svm import pandas as pd from sklearn.ensemble import RandomForestClassifier #load data iris = load_iris() Y = iris.target X = iris.data X_train,X_test,Y_train,Y_test = train_test_split(X,Y,test_size=0.3) clf = RandomForestClassifier(n_estimators=5) clf.fit(X_train, Y_train) score = clf.score(X_test, Y_test) print("模型测试集准确率为:", score) print('特征重要程度: ',clf.feature_importances_)

Work Hard
But do not forget to enjoy life😀
本文来自博客园,作者:YuhangLiuCE,转载请注明原文链接:https://www.cnblogs.com/YuhangLiuCE/p/17978556

