《机器学习》第一次作业——第一至三章学习记录和心得
《机器学习》第一次作业——第一至三章学习记录和心得
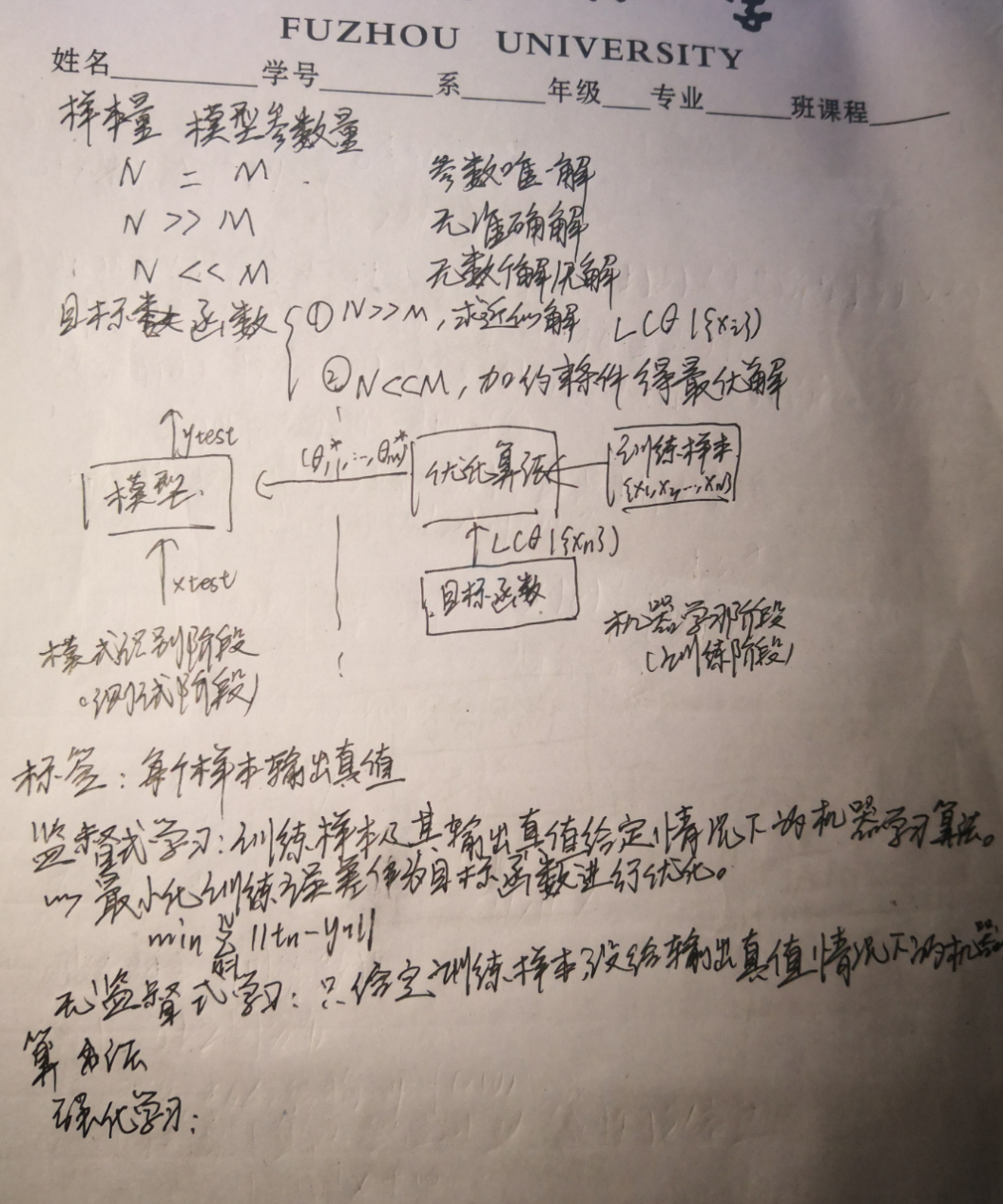
orz懒人直接上图了,真的好多作业好多考试啊啊啊啊啊啊啊啊啊啊








关于讨论区作业
机器学习的作业先做完了,所以可以参考这篇博客,看得出来实验班的大哥哥大姐姐们还是勤奋的,居然有40+访问量了……
1.复现MICD分类器的源码
请根据第二章的理论知识,尝试用Python、MATLAB等常见语言复现MICD分类器。
可以在讨论区内跟大家分享一下自己的代码
from sklearn import datasets
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal as gaussian_cal
#使用np的数组方便向量运算
def getIrisLinear(data,iris_type,flag):
data_linear = [data[i] for i in range(len(data)) if iris_type[i]!=flag]
iris_type_linear = [iris_type[i] for i in range(len(iris_type)) if iris_type[i]!=flag]
return np.asarray(data_linear,dtype="float64"),np.asarray(iris_type_linear,dtype="float64")
def hold_out_partition(data_linear,iris_type_linear):
import random
train_data = []
train_type = []
test_data = []
test_type = []
first_cur = []
second_cur = []
for i in range(len(data_linear)):
if iris_type_linear[i] == 0:
first_cur.append(i)
else:
second_cur.append(i)
k = len(first_cur)-1
#七三开训练集和测试集
train_size = int(len(first_cur) * 7 / 10)
test_size = int(len(first_cur) * 3 / 10)
for i in range(0,train_size):
cur = random.randint(0,k)
train_data.append(data_linear[first_cur[cur]])
train_type.append(iris_type_linear[first_cur[cur]])
k = k - 1
first_cur.remove(first_cur[cur])
for i in range(len(first_cur)):
test_data.append(data_linear[first_cur[i]])
test_type.append(iris_type_linear[first_cur[i]])
k = len(second_cur)-1
train_size = int(len(second_cur) * 7 / 10)
test_size = int(len(second_cur) * 3 / 10)
for i in range(0, train_size):
cur = random.randint(0, k)
train_data.append(data_linear[second_cur[cur]])
train_type.append(iris_type_linear[second_cur[cur]])
k = k - 1
second_cur.remove(second_cur[cur])
for i in range(len(second_cur)):
test_data.append(data_linear[second_cur[i]])
test_type.append(iris_type_linear[second_cur[i]])
return np.asarray(train_data,dtype="float64"),np.asarray(train_type,dtype="int16"),np.asarray(test_data,dtype="float64"),np.asarray(test_type,dtype="int16")
def MED_linear_classification(data,iris_type,t,f,flag):
data_linear,iris_type_linear=getIrisLinear(data,iris_type,flag)
train_data,train_type,test_data,test_type = hold_out_partition(data_linear,iris_type_linear)
c1 = []
c2 = []
n1=0
n2=0
#计算均值
for i in range(len(train_data)):
if train_type[i] == 1:
n1+=1
c1.append(train_data[i])
else:
n2+=1
c2.append(train_data[i])
c1 = np.asarray(c1)
c2 = np.asarray(c2)
z1 = c1.sum(axis=0)/n1
z2 = c2.sum(axis=0)/n2
test_result = []
for i in range(len(test_data)):
result = np.dot(z2-z1,test_data[i]-(z1+z2)/2)
test_result.append(np.sign(result))
test_result = np.array(test_result)
TP = 0
FN = 0
TN = 0
FP = 0
for i in range(len(test_result)):
if(test_result[i]>=0 and test_type[i]==t):
TP+=1
elif(test_result[i]>=0 and test_type[i]==f):
FN+=1
elif(test_result[i]<0 and test_type[i]==t):
FP+=1
elif(test_result[i]<0 and test_type[i]==f):
TN+=1
Recall = TP/(TP+FN)
Precision = TP/(TP+FP)
print("Recall= %f"% Recall)
print("Specify= %f"% (TN/(TN+FP)))
print("Precision= %f"% Precision)
print("F1 Score= %f"% (2*Recall*Precision/(Recall+Precision)))
#开始画图
xx = [[0, 1, 2], [1, 2, 3], [0, 2, 3], [0, 1, 3]]
iris_name =['setosa','vesicolor','virginica']
iris_color = ['r','g','b']
iris_icon = ['o','x','^']
fig = plt.figure(figsize=(20, 20))
feature = ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
for i in range(4):
ax = fig.add_subplot(221 + i, projection="3d")
X = np.arange(test_data.min(axis=0)[xx[i][0]],test_data.max(axis=0)[xx[i][0]],1)
Y = np.arange(test_data.min(axis=0)[xx[i][1]],test_data.max(axis=0)[xx[i][1]],1)
X,Y = np.meshgrid(X,Y)
m1 = [z1[xx[i][0]],z1[xx[i][1]],z1[xx[i][2]]]
m2 = [z2[xx[i][0]], z2[xx[i][1]], z2[xx[i][2]]]
m1 = np.array(m1)
m2 = np.array(m2)
m = m2-m1
#公式化简可得
Z = (np.dot(m,(m1+m2)/2)-m[0]*X-m[1]*Y)/m[2]
ax.scatter(test_data[test_result >= 0, xx[i][0]], test_data[test_result>=0, xx[i][1]], test_data[test_result >= 0, xx[i][2]],
c=iris_color[t], marker=iris_icon[t], label=iris_name[t])
ax.scatter(test_data[test_result < 0, xx[i][0]], test_data[test_result < 0, xx[i][1]],
test_data[test_result < 0, xx[i][2]],
c=iris_color[f], marker=iris_icon[f], label=iris_name[f])
ax.set_zlabel(feature[xx[i][2]])
ax.set_xlabel(feature[xx[i][0]])
ax.set_ylabel(feature[xx[i][1]])
ax.plot_surface(X,Y,Z,alpha=0.4)
plt.legend(loc=0)
plt.show()
def whiten_feature(data):
Ex = np.cov(data,rowvar=False)#这个一定要加……因为我们计算的是特征的协方差
a,w1 = np.linalg.eig(Ex)
w1 = np.real(w1)
module = []
for i in range(w1.shape[1]):
sum = 0
for j in range(w1.shape[0]):
sum += w1[i][j]**2
module.append(sum**0.5)
module = np.asarray(module,dtype="float64")
w1 = w1/module
a = np.real(a)
a=a**(-0.5)
w2 = np.diag(a)
w = np.dot(w2,w1.transpose())
for i in range(w.shape[0]):
for j in range(w.shape[1]):
if np.isnan(w[i][j]):
w[i][j]=0
#print(w)
return np.dot(data,w)
def show_whiten_3D(data,iris_type):
whiten_array = whiten_feature(data)
show_3D(whiten_array,iris_type)
2.复现MICD分类器的源码
请根据第二章的理论知识,尝试用Python、MATLAB等常见语言复现MICD分类器。
可以在讨论区内跟大家分享一下自己的代码
from sklearn import datasets
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal as gaussian_cal
#使用np的数组方便向量运算
def getIrisLinear(data,iris_type,flag):
data_linear = [data[i] for i in range(len(data)) if iris_type[i]!=flag]
iris_type_linear = [iris_type[i] for i in range(len(iris_type)) if iris_type[i]!=flag]
return np.asarray(data_linear,dtype="float64"),np.asarray(iris_type_linear,dtype="float64")
def hold_out_partition(data_linear,iris_type_linear):
import random
train_data = []
train_type = []
test_data = []
test_type = []
first_cur = []
second_cur = []
for i in range(len(data_linear)):
if iris_type_linear[i] == 0:
first_cur.append(i)
else:
second_cur.append(i)
k = len(first_cur)-1
#七三开训练集和测试集
train_size = int(len(first_cur) * 7 / 10)
test_size = int(len(first_cur) * 3 / 10)
for i in range(0,train_size):
cur = random.randint(0,k)
train_data.append(data_linear[first_cur[cur]])
train_type.append(iris_type_linear[first_cur[cur]])
k = k - 1
first_cur.remove(first_cur[cur])
for i in range(len(first_cur)):
test_data.append(data_linear[first_cur[i]])
test_type.append(iris_type_linear[first_cur[i]])
k = len(second_cur)-1
train_size = int(len(second_cur) * 7 / 10)
test_size = int(len(second_cur) * 3 / 10)
for i in range(0, train_size):
cur = random.randint(0, k)
train_data.append(data_linear[second_cur[cur]])
train_type.append(iris_type_linear[second_cur[cur]])
k = k - 1
second_cur.remove(second_cur[cur])
for i in range(len(second_cur)):
test_data.append(data_linear[second_cur[i]])
test_type.append(iris_type_linear[second_cur[i]])
return np.asarray(train_data,dtype="float64"),np.asarray(train_type,dtype="int16"),np.asarray(test_data,dtype="float64"),np.asarray(test_type,dtype="int16")
def MED_linear_classification(data,iris_type,t,f,flag):
data_linear,iris_type_linear=getIrisLinear(data,iris_type,flag)
train_data,train_type,test_data,test_type = hold_out_partition(data_linear,iris_type_linear)
c1 = []
c2 = []
n1=0
n2=0
#计算均值
for i in range(len(train_data)):
if train_type[i] == 1:
n1+=1
c1.append(train_data[i])
else:
n2+=1
c2.append(train_data[i])
c1 = np.asarray(c1)
c2 = np.asarray(c2)
z1 = c1.sum(axis=0)/n1
z2 = c2.sum(axis=0)/n2
test_result = []
for i in range(len(test_data)):
result = np.dot(z2-z1,test_data[i]-(z1+z2)/2)
test_result.append(np.sign(result))
test_result = np.array(test_result)
TP = 0
FN = 0
TN = 0
FP = 0
for i in range(len(test_result)):
if(test_result[i]>=0 and test_type[i]==t):
TP+=1
elif(test_result[i]>=0 and test_type[i]==f):
FN+=1
elif(test_result[i]<0 and test_type[i]==t):
FP+=1
elif(test_result[i]<0 and test_type[i]==f):
TN+=1
Recall = TP/(TP+FN)
Precision = TP/(TP+FP)
print("Recall= %f"% Recall)
print("Specify= %f"% (TN/(TN+FP)))
print("Precision= %f"% Precision)
print("F1 Score= %f"% (2*Recall*Precision/(Recall+Precision)))
#开始画图
xx = [[0, 1, 2], [1, 2, 3], [0, 2, 3], [0, 1, 3]]
iris_name =['setosa','vesicolor','virginica']
iris_color = ['r','g','b']
iris_icon = ['o','x','^']
fig = plt.figure(figsize=(20, 20))
feature = ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
for i in range(4):
ax = fig.add_subplot(221 + i, projection="3d")
X = np.arange(test_data.min(axis=0)[xx[i][0]],test_data.max(axis=0)[xx[i][0]],1)
Y = np.arange(test_data.min(axis=0)[xx[i][1]],test_data.max(axis=0)[xx[i][1]],1)
X,Y = np.meshgrid(X,Y)
m1 = [z1[xx[i][0]],z1[xx[i][1]],z1[xx[i][2]]]
m2 = [z2[xx[i][0]], z2[xx[i][1]], z2[xx[i][2]]]
m1 = np.array(m1)
m2 = np.array(m2)
m = m2-m1
#公式化简可得
Z = (np.dot(m,(m1+m2)/2)-m[0]*X-m[1]*Y)/m[2]
ax.scatter(test_data[test_result >= 0, xx[i][0]], test_data[test_result>=0, xx[i][1]], test_data[test_result >= 0, xx[i][2]],
c=iris_color[t], marker=iris_icon[t], label=iris_name[t])
ax.scatter(test_data[test_result < 0, xx[i][0]], test_data[test_result < 0, xx[i][1]],
test_data[test_result < 0, xx[i][2]],
c=iris_color[f], marker=iris_icon[f], label=iris_name[f])
ax.set_zlabel(feature[xx[i][2]])
ax.set_xlabel(feature[xx[i][0]])
ax.set_ylabel(feature[xx[i][1]])
ax.plot_surface(X,Y,Z,alpha=0.4)
plt.legend(loc=0)
plt.show()
def whiten_feature(data):
Ex = np.cov(data,rowvar=False)#这个一定要加……因为我们计算的是特征的协方差
a,w1 = np.linalg.eig(Ex)
w1 = np.real(w1)
module = []
for i in range(w1.shape[1]):
sum = 0
for j in range(w1.shape[0]):
sum += w1[i][j]**2
module.append(sum**0.5)
module = np.asarray(module,dtype="float64")
w1 = w1/module
a = np.real(a)
a=a**(-0.5)
w2 = np.diag(a)
w = np.dot(w2,w1.transpose())
for i in range(w.shape[0]):
for j in range(w.shape[1]):
if np.isnan(w[i][j]):
w[i][j]=0
#print(w)
return np.dot(data,w)
def show_whiten_3D(data,iris_type):
whiten_array = whiten_feature(data)
show_3D(whiten_array,iris_type)


 浙公网安备 33010602011771号
浙公网安备 33010602011771号