《机器学习》上机实践(1)
《机器学习》上机实践(1)
代码有参考往年学长的博客orz:转这里麻了麻了,很认真的看了好久才懂咋实现的
题目
-
Iris数据集已与常见的机器学习工具集成,请查阅资料找出MATLAB平台或Python平台加载内置Iris数据集方法,并简要描述该数据集结构。
核心代码如下:
from sklearn import datasets
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal as gaussian_cal
Iris = datasets.load_iris()
数据结构如下:
很明显是一个字典
{
data:...
target:...
target_names:...
...
}
比较主要的就这三个关键字,data是每个数据的特征数组,target是每个数据的归类,target_names是每类数据的名字
-
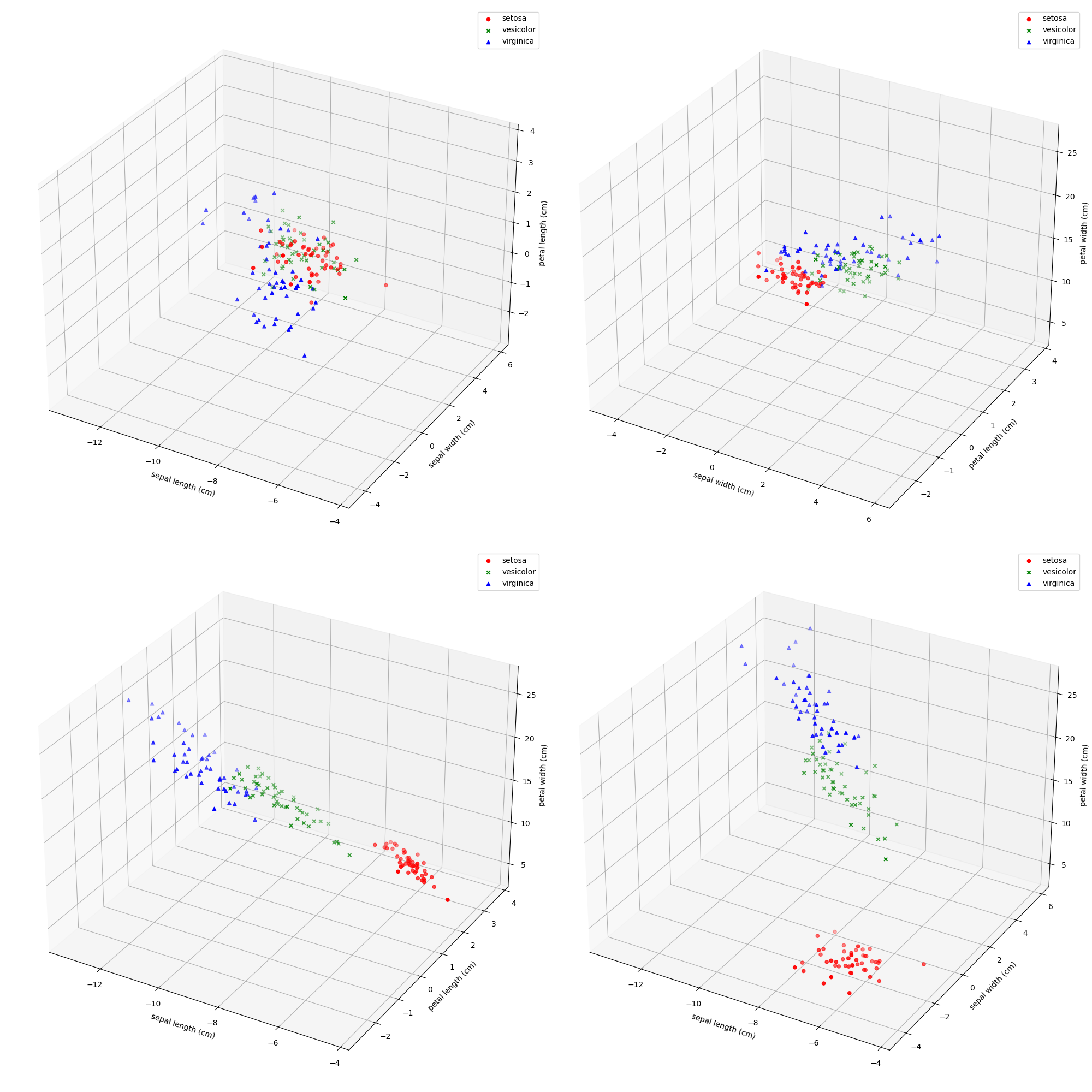
Iris数据集中有一个种类与另外两个类是线性可分的,其余两个类是线性不可分的。请你通过数据可视化的方法找出该线性可分类并给出判断依据。
orzPCA降维可以降一维画四张图挺不错的,但是我还是老实研究了一波三维图
很明显setosa和另外两个是线性可分的,剩下两类是线性不可分的

核心代码
from sklearn import datasets
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal as gaussian_cal
def show_3D(data,iris_type):
xx = [[0, 1, 2], [1, 2, 3], [0, 2, 3], [0, 1, 3]]
fig = plt.figure(figsize=(20, 20))
feature = ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
for i in range(4):
ax = fig.add_subplot(221 + i, projection="3d")
ax.scatter(data[iris_type == 0, xx[i][0]], data[iris_type == 0, xx[i][1]], data[iris_type == 0, xx[i][2]],
c='r', marker='o', label='setosa')
ax.scatter(data[iris_type == 1, xx[i][0]], data[iris_type == 1, xx[i][1]], data[iris_type == 1, xx[i][2]],
c='g', marker='x',
label='vesicolor')
ax.scatter(data[iris_type == 2, xx[i][0]], data[iris_type == 2, xx[i][1]], data[iris_type == 2, xx[i][2]],
c='b', marker='^',
label='virginica')
ax.set_zlabel(feature[xx[i][2]])
ax.set_xlabel(feature[xx[i][0]])
ax.set_ylabel(feature[xx[i][1]])
plt.legend(loc=0)
plt.show()
-
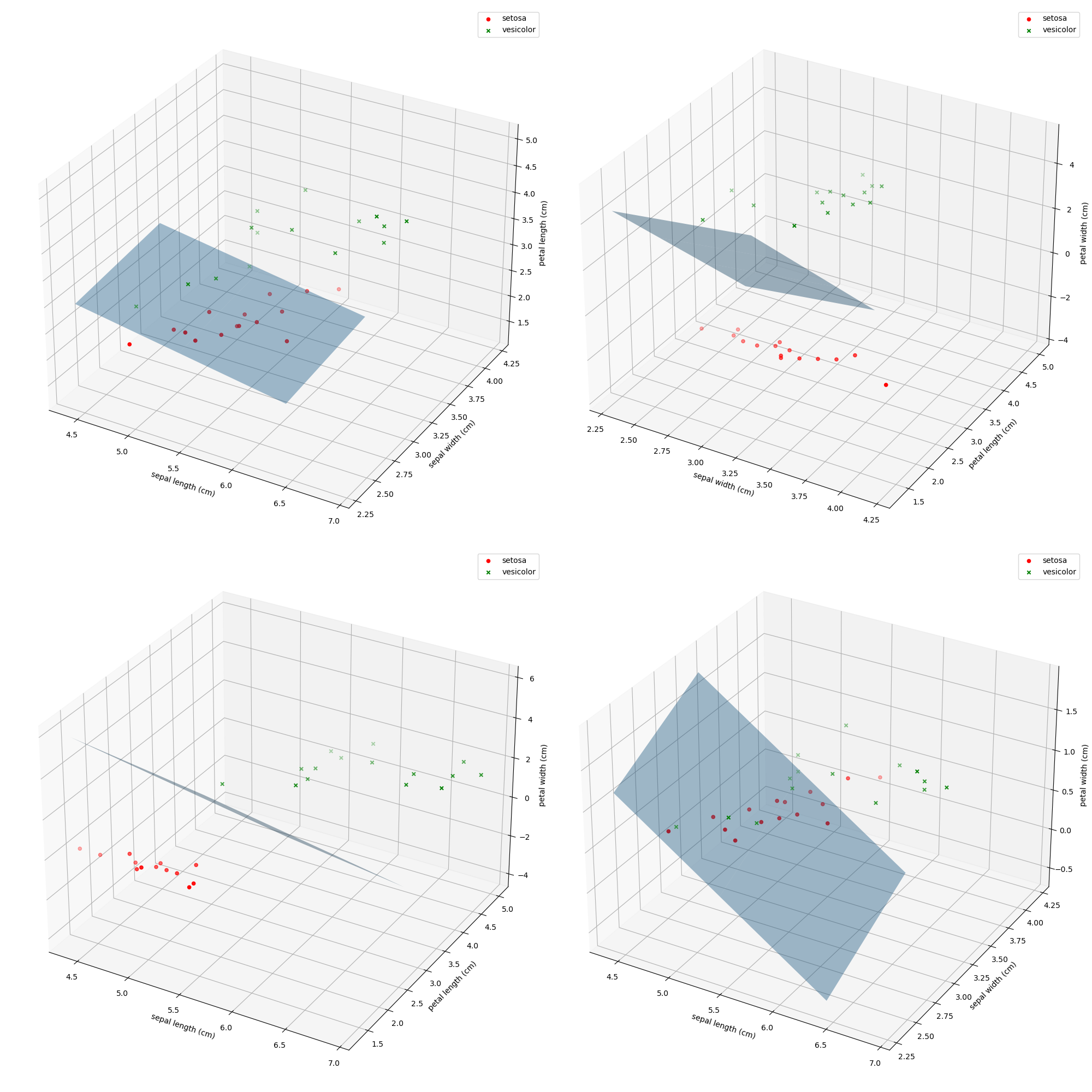
去除Iris数据集中线性不可分的类中最后一个,余下的两个线性可分的类构成的数据集命令为Iris_linear,请使用留出法将Iris_linear数据集按7:3分为训练集与测试集,并使用训练集训练一个MED分类器,在测试集上测试训练好的分类器的性能,给出《模式识别与机器学习-评估方法与性能指标》中所有量化指标并可视化分类结果。
留出法这边采用的是等比例每类里随机取数
def hold_out_partition(data_linear,iris_type_linear):
import random
train_data = []
train_type = []
test_data = []
test_type = []
first_cur = []
second_cur = []
for i in range(len(data_linear)):
if iris_type_linear[i] == 0:
first_cur.append(i)
else:
second_cur.append(i)
k = len(first_cur)-1
#七三开训练集和测试集
train_size = int(len(first_cur) * 7 / 10)
test_size = int(len(first_cur) * 3 / 10)
for i in range(0,train_size):
cur = random.randint(0,k)
train_data.append(data_linear[first_cur[cur]])
train_type.append(iris_type_linear[first_cur[cur]])
k = k - 1
first_cur.remove(first_cur[cur])
for i in range(len(first_cur)):
test_data.append(data_linear[first_cur[i]])
test_type.append(iris_type_linear[first_cur[i]])
k = len(second_cur)-1
train_size = int(len(second_cur) * 7 / 10)
test_size = int(len(second_cur) * 3 / 10)
for i in range(0, train_size):
cur = random.randint(0, k)
train_data.append(data_linear[second_cur[cur]])
train_type.append(iris_type_linear[second_cur[cur]])
k = k - 1
second_cur.remove(second_cur[cur])
for i in range(len(second_cur)):
test_data.append(data_linear[second_cur[i]])
test_type.append(iris_type_linear[second_cur[i]])
return np.asarray(train_data,dtype="float64"),np.asarray(train_type,dtype="int16"),np.asarray(test_data,dtype="float64"),np.asarray(test_type,dtype="int16")
由于是线性可分的,各项指标到达1.0
Recall= 1.000000
Specify= 1.000000
Precision= 1.000000
F1 Score= 1.000000
这边界平面真是给画去世了……得到均值两点后就有法向量和平面上一点,平面方程就有了
from sklearn import datasets
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal as gaussian_cal
def MED_linear_classification(data,iris_type,t,f,flag):
data_linear,iris_type_linear=getIrisLinear(data,iris_type,flag)
train_data,train_type,test_data,test_type = hold_out_partition(data_linear,iris_type_linear)
c1 = []
c2 = []
n1=0
n2=0
#计算均值
for i in range(len(train_data)):
if train_type[i] == 1:
n1+=1
c1.append(train_data[i])
else:
n2+=1
c2.append(train_data[i])
c1 = np.asarray(c1)
c2 = np.asarray(c2)
z1 = c1.sum(axis=0)/n1
z2 = c2.sum(axis=0)/n2
test_result = []
for i in range(len(test_data)):
result = np.dot(z2-z1,test_data[i]-(z1+z2)/2)
test_result.append(np.sign(result))
test_result = np.array(test_result)
TP = 0
FN = 0
TN = 0
FP = 0
for i in range(len(test_result)):
if(test_result[i]>=0 and test_type[i]==t):
TP+=1
elif(test_result[i]>=0 and test_type[i]==f):
FN+=1
elif(test_result[i]<0 and test_type[i]==t):
FP+=1
elif(test_result[i]<0 and test_type[i]==f):
TN+=1
Recall = TP/(TP+FN)
Precision = TP/(TP+FP)
print("Recall= %f"% Recall)
print("Specify= %f"% (TN/(TN+FP)))
print("Precision= %f"% Precision)
print("F1 Score= %f"% (2*Recall*Precision/(Recall+Precision)))
#开始画图
xx = [[0, 1, 2], [1, 2, 3], [0, 2, 3], [0, 1, 3]]
iris_name =['setosa','vesicolor','virginica']
iris_color = ['r','g','b']
iris_icon = ['o','x','^']
fig = plt.figure(figsize=(20, 20))
feature = ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
for i in range(4):
ax = fig.add_subplot(221 + i, projection="3d")
X = np.arange(test_data.min(axis=0)[xx[i][0]],test_data.max(axis=0)[xx[i][0]],1)
Y = np.arange(test_data.min(axis=0)[xx[i][1]],test_data.max(axis=0)[xx[i][1]],1)
X,Y = np.meshgrid(X,Y)
m1 = [z1[xx[i][0]],z1[xx[i][1]],z1[xx[i][2]]]
m2 = [z2[xx[i][0]], z2[xx[i][1]], z2[xx[i][2]]]
m1 = np.array(m1)
m2 = np.array(m2)
m = m2-m1
#公式化简可得
Z = (np.dot(m,(m1+m2)/2)-m[0]*X-m[1]*Y)/m[2]
ax.scatter(test_data[test_result >= 0, xx[i][0]], test_data[test_result>=0, xx[i][1]], test_data[test_result >= 0, xx[i][2]],
c=iris_color[t], marker=iris_icon[t], label=iris_name[t])
ax.scatter(test_data[test_result < 0, xx[i][0]], test_data[test_result < 0, xx[i][1]],
test_data[test_result < 0, xx[i][2]],
c=iris_color[f], marker=iris_icon[f], label=iris_name[f])
ax.set_zlabel(feature[xx[i][2]])
ax.set_xlabel(feature[xx[i][0]])
ax.set_ylabel(feature[xx[i][1]])
ax.plot_surface(X,Y,Z,alpha=0.4)
plt.legend(loc=0)
plt.show()
-
将Iris数据集白化,可视化白化结果并于原始可视化结果比较,讨论白化的作用。
只能说numpy真的很香,能支持向量运算,白化后数据确实直观很多
代码如下:
def whiten_feature(data):
Ex = np.cov(data,rowvar=False)#这个一定要加……因为我们计算的是特征的协方差
a,w1 = np.linalg.eig(Ex)
w1 = np.real(w1)
module = []
for i in range(w1.shape[1]):
sum = 0
for j in range(w1.shape[0]):
sum += w1[i][j]**2
module.append(sum**0.5)
module = np.asarray(module,dtype="float64")
w1 = w1/module
a = np.real(a)
a=a**(-0.5)
w2 = np.diag(a)
w = np.dot(w2,w1.transpose())
for i in range(w.shape[0]):
for j in range(w.shape[1]):
if np.isnan(w[i][j]):
w[i][j]=0
#print(w)
return np.dot(data,w)
-
去除Iris数据集中线性可分的类,余下的两个线性不可分的类构成的数据集命令为Iris_nonlinear,请使用留出法将Iris_nonlinear数据集按7:3分为训练集与测试集,并使用训练集训练一个MED分类器,在测试集上测试训练好的分类器的性能,给出《模式识别与机器学习-评估方法与性能指标》中所有量化指标并可视化分类结果。讨论本题结果与3题结果的差异。
同样的代码不一样的数据集,由于随机性以及数据的线性不可分的原因,各项指标每次测量都不一样,有的时候会有除0风险orz,对边界的确定影响很大
Recall= 0.066667
Specify= 0.066667
Precision= 0.066667
F1 Score= 0.066667

-
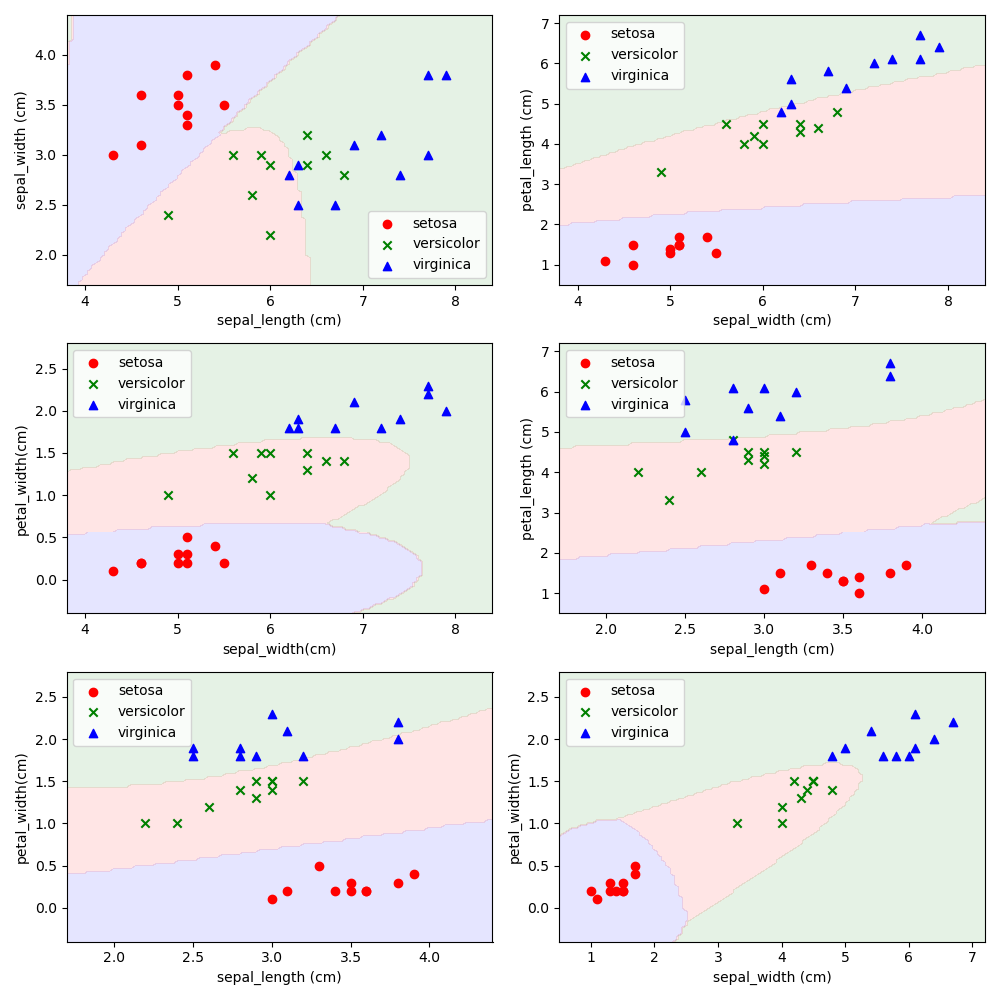
请使用5折交叉验证为Iris数据集训练一个多分类的贝叶斯分类器。给出平均Accuracy,并可视化实验结果。与第3题和第5题结果做比较,讨论贝叶斯分类器的优劣。
只能说,做麻了,效果确实比MED好
Accuracy = 0.9733333333333334,基本在这个值上下浮动,因为k折验证也是随机取点
代码:
from sklearn import datasets
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal as gaussian_cal
def k_split(data,iris_type,num):
import random
testSet = []
testType = []
first_cur = []
second_cur = []
third_cur = []
for i in range(len(iris_type)):
if iris_type[i] == 0:
first_cur.append(i)
elif iris_type[i] == 1:
second_cur.append(i)
else:
third_cur.append(i)
match_size = int(len(first_cur)/num)
size = len(first_cur)-1
train_data = []
train_type = []
for i in range(num):
k = match_size
train_data = []
train_type = []
for j in range(match_size):
cur = random.randint(0, size)
train_data.append(data[first_cur[cur]])
train_type.append(iris_type[first_cur[cur]])
first_cur.remove(first_cur[cur])
cur = random.randint(0, size)
train_data.append(data[second_cur[cur]])
train_type.append(iris_type[second_cur[cur]])
second_cur.remove(second_cur[cur])
cur = random.randint(0, size)
train_data.append(data[third_cur[cur]])
train_type.append(iris_type[third_cur[cur]])
third_cur.remove(third_cur[cur])
size = size-1
testSet.append(train_data)
testType.append(train_type)
return np.asarray(testSet),np.asarray(testType)
class Bayes_Parameter():
def __init__(self,mean,cov,type):
self.mean = mean
self.cov = cov
self.type = type
class Bayes_Classifier():
#必须存入k-1个训练集的每个高斯分布
def __init__(self):
self.parameters=[]
def train(self,data,iris_type):
for type in set(iris_type):
selected = iris_type==type
select_data = data[selected]
mean = np.mean(select_data,axis=0)
cov = np.cov(select_data.transpose())
self.parameters.append(Bayes_Parameter(mean,cov,type))
def predict(self,data):
result = -1
probability = 0
for parameter in self.parameters:
temp = gaussian_cal.pdf(data,parameter.mean,parameter.cov)
if temp > probability:
probability = temp
result = parameter.type
return result
def Bayes_Classification_K_split(data,iris_type,num):
train_dataset,train_typeset = k_split(data,iris_type,num)
accuracy = 0
best_result = []
best_train_data = []
best_train_type = []
best_test_data = []
best_test_type = []
max_accuracy = 0
for i in range(num):
data_num = 0
type_num = 0
train_data = []
train_type = []
for j in range(num):
if i != j:
if data_num*type_num == 0:
train_data = train_dataset[j]
train_type = train_typeset[j]
data_num+=1
type_num+=1
else:
train_data = np.concatenate((train_data,train_dataset[j]),axis=0)
train_type = np.concatenate((train_type,train_typeset[j]),axis=0)
Bayes_classifier = Bayes_Classifier()
Bayes_classifier.train(train_data,train_type)
predict_result = [Bayes_classifier.predict(x) for x in train_dataset[i]]
right = 0
all = 0
for j in range(len(predict_result)):
if predict_result[j] == train_typeset[i][j]:
right+=1
all+=1
tempaccuracy = right/all
if tempaccuracy > max_accuracy:
max_accuracy = tempaccuracy
best_train_data = train_data
best_train_type = train_type
best_test_data = train_dataset[i]
best_test_type = train_typeset[i]
best_result = np.asarray(predict_result,dtype="int")
accuracy+=tempaccuracy
show_2D(best_train_data,best_train_type,best_test_data,best_test_type,best_result)
return accuracy/5
def show_2D(train_data,train_type,test_data,test_type,result):
import math
fig = plt.figure(figsize=(10,10))
xx = [[0,1],[0,2],[0,3],[1,2],[1,3],[2,3]]
yy = [["sepal_length (cm)", "sepal_width (cm)"],
["sepal_width (cm)", "petal_length (cm)"],
["sepal_width(cm)", "petal_width(cm)"],
["sepal_length (cm)", "petal_length (cm)"],
["sepal_length (cm)", "petal_width(cm)"],
["sepal_width (cm)", "petal_width(cm)"]]
for i in range(6):
ax = fig.add_subplot(321+i)
x_max,x_min = test_data.max(axis=0)[xx[i][0]]+0.5,test_data.min(axis=0)[xx[i][0]]-0.5
y_max,y_min = test_data.max(axis=0)[xx[i][1]]+0.5,test_data.min(axis=0)[xx[i][1]]-0.5
xlist = np.linspace(x_min, x_max, 100)
ylist = np.linspace(y_min, y_max, 100)
X, Y = np.meshgrid(xlist,ylist)
bc = Bayes_Classifier()
bc.train(train_data[:,xx[i]],train_type)
xy = [np.array([xx,yy]).reshape(1,-1 ) for xx,yy in zip(np.ravel(X),np.ravel(Y))]
zz = np.array([bc.predict(x) for x in xy])
Z = zz.reshape(X.shape)
plt.contourf(X,Y,Z,2,alpha=.1,colors=('blue','red','green'))
ax.scatter(test_data[result==0,xx[i][0]],test_data[result==0,xx[i][1]],c='r',marker='o',label='setosa')
ax.scatter(test_data[result == 1, xx[i][0]], test_data[result == 1, xx[i][1]], c='g', marker='x',
label='versicolor')
ax.scatter(test_data[result == 2, xx[i][0]], test_data[result == 2, xx[i][1]], c='b', marker='^', label='virginica')
ax.set_xlabel(yy[i][0])
ax.set_ylabel(yy[i][1])
ax.legend(loc=0)
plt.show()
感想
其实分类海星,就是作图实在是……代码实现很麻,主要是对numpy,sklearn,scipy,matplotlib这些库不是很熟吧,除了学长的博客也查了很多其他这些库的内容,包括如何用python画三维图,sklearn计算高斯分布概率啥的,对我个人来说锻炼还是很多的吧,很久没写代码了orz


 浙公网安备 33010602011771号
浙公网安备 33010602011771号