聚类指标
1 简介
之前接触过一段时间聚类算法,这里记录一下在聚类中常用的评价指标,并给出相应的代码。假设我们对10个对象进行聚类,这10个对象的原始标签为[0, 0, 0, 1, 1, 1, 2, 2, 3, 3],那么预测标签[1, 2, 0, 2, 2, 2, 3, 3, 3, 0]和[2, 3, 1, 3, 3, 3, 0, 0, 0, 1]得到的指标应该一样(对于同样的聚类结果,即便给各个簇分配不同ID,其得到的指标结果应该一样)。
2 常用聚类指标
假设对N个点进行聚类,N个点之间可以形成\(\frac{N \times (N-1)}{2}\)条连边。如下图所示,我们可以对这些连边进行分类:
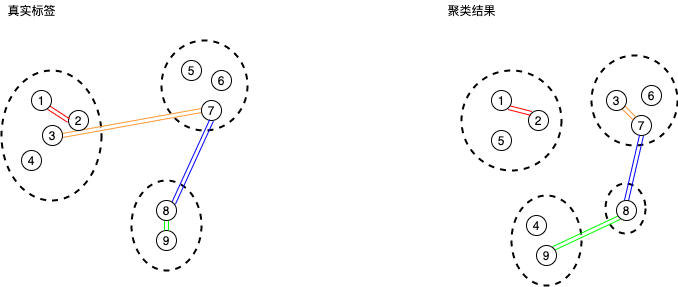
第一类(红色):在真实标签和聚类结果下都属于同一个cluster,TP。
第二类(绿色):在真实标签下属于一个cluster,在聚类结果中不属于一个cluster,FP。
第三类(橙色):在真实标签下不属于一个cluster,在聚类结果下属于一个cluster,FN。
第四类(蓝色):在真实标签和聚类结果下都不属于一个clusger,TN。
根据sklearn.metrics.cluster.fowlkes_mallows_score,这里的FP和FN定义好像和很多资料中的相反。
2.1 FMI
Fowlkes-Mallows index (FMI)定义为precision和recall的几何平均
\(FMI = \sqrt{\frac{TP \times TP}{(TP + FN) \times (TP + FP)}}\)
def fowlkes_mallows_score(labels_true, labels_pred, *, sparse=False):
"""Measure the similarity of two clusterings of a set of points.
.. versionadded:: 0.18
The Fowlkes-Mallows index (FMI) is defined as the geometric mean between of
the precision and recall::
FMI = TP / sqrt((TP + FP) * (TP + FN))
Where ``TP`` is the number of **True Positive** (i.e. the number of pair of
points that belongs in the same clusters in both ``labels_true`` and
``labels_pred``), ``FP`` is the number of **False Positive** (i.e. the
number of pair of points that belongs in the same clusters in
``labels_true`` and not in ``labels_pred``) and ``FN`` is the number of
**False Negative** (i.e the number of pair of points that belongs in the
same clusters in ``labels_pred`` and not in ``labels_True``).
The score ranges from 0 to 1. A high value indicates a good similarity
between two clusters.
Read more in the :ref:`User Guide <fowlkes_mallows_scores>`.
Parameters
----------
labels_true : int array, shape = (``n_samples``,)
A clustering of the data into disjoint subsets.
labels_pred : array, shape = (``n_samples``, )
A clustering of the data into disjoint subsets.
sparse : bool, default=False
Compute contingency matrix internally with sparse matrix.
Returns
-------
score : float
The resulting Fowlkes-Mallows score.
Examples
--------
Perfect labelings are both homogeneous and complete, hence have
score 1.0::
>>> from sklearn.metrics.cluster import fowlkes_mallows_score
>>> fowlkes_mallows_score([0, 0, 1, 1], [0, 0, 1, 1])
1.0
>>> fowlkes_mallows_score([0, 0, 1, 1], [1, 1, 0, 0])
1.0
If classes members are completely split across different clusters,
the assignment is totally random, hence the FMI is null::
>>> fowlkes_mallows_score([0, 0, 0, 0], [0, 1, 2, 3])
0.0
References
----------
.. [1] `E. B. Fowkles and C. L. Mallows, 1983. "A method for comparing two
hierarchical clusterings". Journal of the American Statistical
Association
<http://wildfire.stat.ucla.edu/pdflibrary/fowlkes.pdf>`_
.. [2] `Wikipedia entry for the Fowlkes-Mallows Index
<https://en.wikipedia.org/wiki/Fowlkes-Mallows_index>`_
"""
labels_true, labels_pred = check_clusterings(labels_true, labels_pred)
n_samples, = labels_true.shape
c = contingency_matrix(labels_true, labels_pred,
sparse=True)
c = c.astype(np.int64, **_astype_copy_false(c))
tk = np.dot(c.data, c.data) - n_samples
pk = np.sum(np.asarray(c.sum(axis=0)).ravel() ** 2) - n_samples
qk = np.sum(np.asarray(c.sum(axis=1)).ravel() ** 2) - n_samples
return np.sqrt(tk / pk) * np.sqrt(tk / qk) if tk != 0. else 0.
2.2 Pair-wise F-Score
Pair-wise Precision和Pair-wise Recall和FMI中的计算一样,只不过不同于FMI求的是precision和recall的几何平均,Pair-wise F-Score计算的是precision和recall的调和平均
2.3 BCubed F-Score
参考Linkage论文,BCubed指标的计算公式:
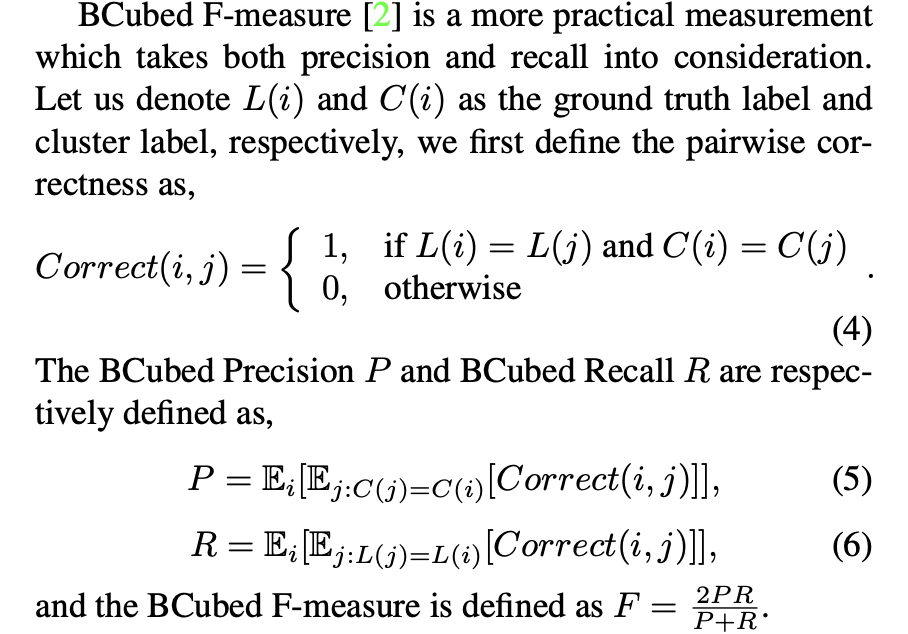
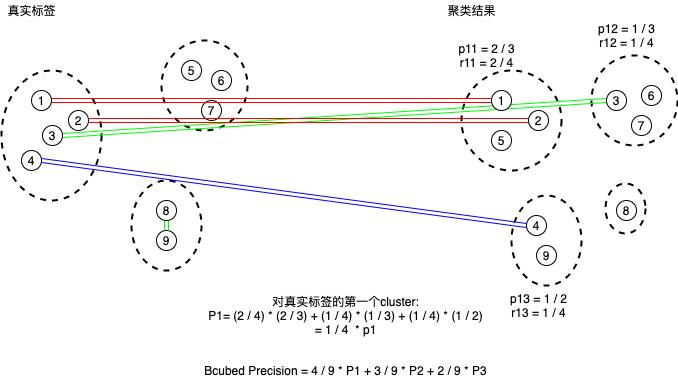
可以用下面这张图来理解计算过程及后面的bcubed函数。
2.4 NMI
NMI: Normalized Mutual Information
对数据\(D=\{x_1, x_2, x_3, ..., x_n\}\)进行聚类,假设通过聚类得到p个cluster,\(C = \{C_1, C_2, ..., C_p\}\),根据标签得到q个cluster,\(C^* = \{C^*_1, C^*_2, ..., C^*_q\}\),则:
\(\begin{aligned} M I\left(C, C^{*}\right) &=\sum_{i=1}^{p} \sum_{j=1}^{q} P\left(C_{i}, C_{j}^{*}\right) \log \frac{P\left(C_{i} \cap C_{j}^{*}\right)}{P\left(C_{i}\right) P\left(C_{j}^{*}\right)} \\ &=\sum_{i=1}^{p} \sum_{j=1}^{q} \frac{\left|C_{i} \cap C_{j}^{*}\right|}{n} \log \frac{n \cdot\left|C_{i} \cap C_{j}^{*}\right|}{\left|C_{i}\right|\left|C_{j}^{*}\right|} \end{aligned}\)
这个公式也可以结合解释BCubed的图示进一步理解。
下面的\(H(C)\)是信息熵的形式,它可以用来表示不确定性度量
\(\begin{aligned} H(C) &=-\sum_{i=1}^{p} P\left(C_{i}\right) \log P\left(C_{i}\right) \\ &=-\sum_{i=1}^{p} \frac{\left|C_{i}\right|}{n} \log \left(\frac{\left|C_{i}\right|}{n}\right) \end{aligned}\)
根据互信息和信息熵的关系,我们有:
\(M I\left(C, C^{*}\right)=H(C)-H\left(C \mid C^{*}\right)\)
如果互信息(mutual information)越大,说明两者的关系越密切(如果\(H(C) = H\left(C \mid C^{*}\right)\),说明了解\(C^{*}\)不能为\(C\)提供任何有用的信息、以减少\(C\)的不确定性)。对于聚类结果,我们期望MI值能更大。但是如果将所有样本各划分成1类,\(MI(C, C^{*})\)会达到最大(类的split严重,虽然每个cluster的纯度很高,但是召回很低),为了避免这一倾向,使用Normalized mutual information,因为划分的cluster过多时,\(H(C)\)(信息熵,不确定性越大,熵越大)会更大。
\(N M I(C,C^{*})=\frac{MI(C, C^{*})}{\sqrt{H(C) H(C^{*})}}\)
3 指标计算代码
from __future__ import division
import numpy as np
from sklearn.metrics.cluster import contigency_matrix, normalized_mutual_info_score
from skleran.metrics improt precision_score, recall_score
def _check(gt_labels, pred_labels):
if gt_labels.ndim != 1:
raise ValueError("gt_labels must be 1D: shape is {}".format(gt_labels.shape))
if pred_labels.ndim != 1:
raise ValueError("pred_labels must be 1D: shape is {}".format(pred_labels.shape))
if pred_labels.shape != gt_labels.shape:
raise ValueError("gt_labels and pred_labels must have same size, got {} and {}".format(gt_labels.shape, pred_labels.shape))
return gt_labels, pred_labels
def _get_lb2idxs(labels):
lb2idxs = {}
for idx, lb in enumerate(labels):
if lb not in lb2idxs:
lb2idxs[lb] = []
lb2idxs[lb].append(idx)
return lb2idxs
def fowlkes_mallows_score(gt_labels, pred_labels, sparse=True):
n_samples, = gt_labels.shape
c = contingency_matrix(gt_labels, pred_labels, sparse=sparse)
tk = np.dot(c.data, c.data) - n_samples
pk = np.sum(np.asarray(c.sum(axis=0)).ravel()**2) - n_samples
qk = np.sum(np.asarray(c.sum(axis=1)).ravel()**2) - n_samples
avg_pre = tk / pk
avg_rec = tk / qk
fscore = 2. * avg_pre * avg_rec / (avg_pre + avg_rec)
return fscore
def pairwise(gt_labels, pred_labels, sparse=True):
_check(gt_labels, pred_labels)
return fowlkes_mallows_score(gt_labels, pred_labels, sparse)
def bcubed(gt_labels, pred_labels):
gt_lb2idxs = _get_lb2idxs(gt_labels)
pred_lb2idxs = _get_lb2idxs(pred_labels)
num_lbs = len(gt_labels.keys())
pre = np.zeros(num_lbs)
rec = np.zeros(num_lbs)
gt_num = np.zeros(num_lbs)
for idx, gt_idxs in enumerate(gt_lb2idxs.values()):
all_pred_lbs = np.unique(pred_labels[gt_idxs])
gt_num[idx] = len(gt_idxs)
for pred_lb in all_pred_lbs:
pred_idxs = pred_lb2idxs[pred_lb]
n = 1. * np.intersect1d(gt_idxs, pred_idxs).size
# 为什么是 n**2,理解一下
# [n1 * (n1 / p1) + n2 * (n2 / p2) + ... + nk * (nk / pk)] / n
# n = n1 + n2 + ... + nk
pre[idx] += n**2 / len(pred_idxs)
rec[idx] += n**2 / gt_num[i]
gt_num = gt_num.sum()
avg_pre = pre.sum() / gt_sum
avg_rec = rec.sum() / gt_sum
return
def nmi(gt_labels, pred_labels):
return normalized_mutual_info_score(pred_labels, gt_labels)
另外一种实现Bcubed的方式:
import numpy as np
from scipy.sparse import coo_matrix
__all__ = ['bcubed', 'conditional_entropy', 'contingency_matrix', 'der',
'goodman_kruskal_tau', 'mutual_information']
EPS = np.finfo(float).eps
def contingency_matrix(ref_labels, sys_labels):
"""Return contingency matrix between ``ref_labels`` and ``sys_labels``."""
ref_classes, ref_class_inds = np.unique(ref_labels, return_inverse=True)
sys_classes, sys_class_inds = np.unique(sys_labels, return_inverse=True)
n_frames = ref_labels.size
# Following works because coo_matrix sums duplicate entries. Is roughly
# twice as fast as np.histogram2d.
cmatrix = coo_matrix(
(np.ones(n_frames), (ref_class_inds, sys_class_inds)),
shape=(ref_classes.size, sys_classes.size),
dtype=np.int)
cmatrix = cmatrix.toarray()
return cmatrix, ref_classes, sys_classes
def bcubed(ref_labels, sys_labels, cm=None):
"""Return B-cubed precision, recall, and F1.
The B-cubed precision of an item is the proportion of items with its
system label that share its reference label (Bagga and Baldwin, 1998).
Similarly, the B-cubed recall of an item is the proportion pf items
with its reference label that share its system label. The overall B-cubed
precision and recall, then, are the means of the precision and recall for
each item.
Parameters
----------
ref_labels : ndarray, (n_frames,)
Reference labels.
sys_labels : ndarray, (n_frames,)
System labels.
cm : ndarray, (n_ref_classes, n_sys_classes)
Contingency matrix between reference and system labelings. If None,
will be computed automatically from ``ref_labels`` and ``sys_labels``.
Otherwise, the given value will be used and ``ref_labels`` and
``sys_labels`` ignored.
(Default: None)
Returns
-------
precision : float
B-cubed precision.
recall : float
B-cubed recall.
f1 : float
B-cubed F1.
References
----------
Bagga, A. andBaldwin, B. (1998). "Algorithmsfor scoring coreference
chains." Proceedings of LREC 1998.
"""
if cm is None:
cm, _, _ = contingency_matrix(ref_labels, sys_labels)
cm = cm.astype('float64')
cm_norm = cm / cm.sum()
precision = np.sum(cm_norm * (cm / cm.sum(axis=0)))
recall = np.sum(cm_norm * (cm / np.expand_dims(cm.sum(axis=1), 1)))
f1 = 2*(precision*recall)/(precision + recall)
return precision, recall, f1



