SAS waterfall瀑布图
第一部分:统一X轴,只用一个标准。DISPLAY = NONE啥也不显示了。COLORBANDS = ODD隔行换色
第二部分:
filledoutlinedmarkers=true markerfillattrs=(color=gold) markeroutlineattrs=(color=black)
marker有边框,边框色和填充色不一致。
第三部分:BANDPLOT是figure中上下两条蓝色虚线。extend = true会在末尾多延长一点点,在survival plot中和STEPPLOT连用时看起来明显。DATALABELFITPOLICY就是如何显示datalabel。
TEXTPLOT就是把把数据以文本形式展现在figure中。
第四部分:
在GitHub看到的例子,分享在这。
data attrmap中的ID VALUE是必须的,ID必须排序。在PROC SGRENDER中 dattrmap=attrmap dattrvar = ""会调用这些自定义的属性
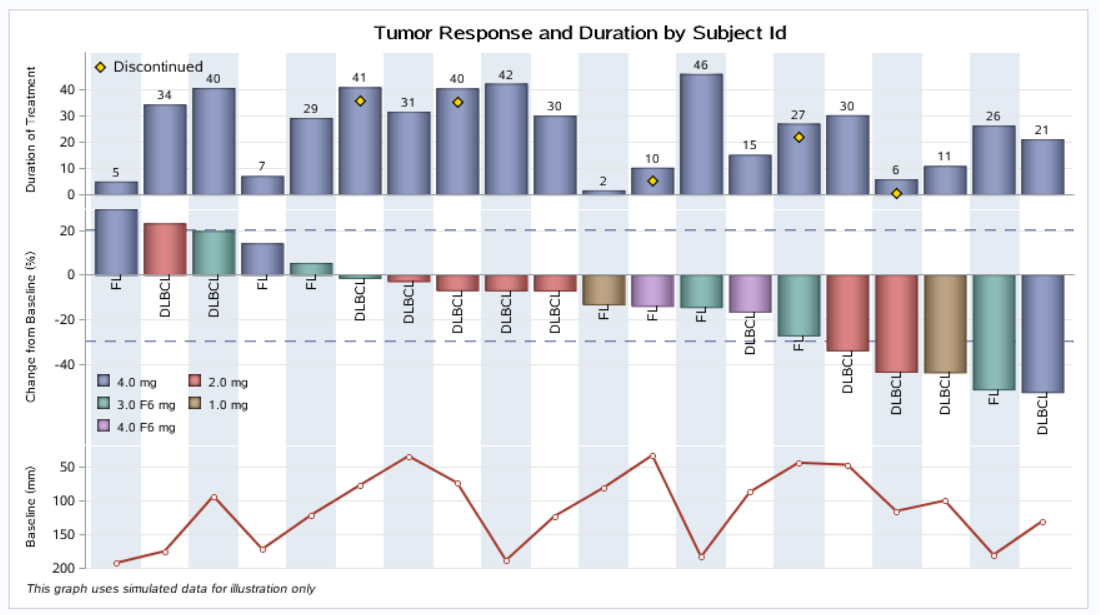
proc format; value res 0='1.0 mg' 1='2.0 mg' 2='3.0 mg' 3='4.0 mg' 4='3.0 F6 mg' 5='4.0 F6 mg'; run; /*--Create some data--*/ data tumor; label Duration='Duration of Treatment' Dropped='Discontinued'; format Drug res.; length Code $6; do i=1 to 20; Response=100*(ranuni(3)-0.7); Duration=50*ranuni(3); Dropped=.; if ranuni(3) > 0.8 then Dropped=duration-5; Drug=floor(6*ranuni(3)); if ranuni(3) > 0.7 then Code='FL'; else Code='DLBCL'; codeloc=ifn(response > 0, 0, response); baseline=20+180*(ranuni(2)); output; end; run; /*--Sort by descending response--*/ proc sort data=tumor out=tumor; by descending Response; run; data tumorsorted; set tumor; j=_n_; run; ods html; /*--Define Attributes map for walls and axes--*/ data attrmap; length ID $ 9 value $10 fillcolor $ 10 show $8; id='Resp'; value='1.0 mg'; fillcolor='white'; show='Attrmap'; output; id='Resp'; value='2.0 mg'; fillcolor='cxffffc0'; show='Attrmap'; output; id='Resp'; value='3.0 mg'; fillcolor='cxf0d0a0'; show='Attrmap'; output; id='Resp'; value='4.0 mg'; fillcolor='orange'; show='Attrmap'; output; id='Resp'; value='3.0 F6 mg'; fillcolor='cxff7f00'; show='Attrmap'; output; id='Resp'; value='4.0 F6 mg'; fillcolor='red'; show='Attrmap'; output; run; /*--WaterFall with Tumor Baseline--*/ proc template; define statgraph Waterfall_Baseline_Series; begingraph / subpixel = on; entrytitle 'Tumor Response and Duration by Subject Id'; entryfootnote halign=left 'This graph uses simulated data for illustration only' / textattrs=(size=7pt style=italic); layout lattice / columndatarange=union rowweights=(0.3 0.45 0.25) rowgutter=0;
*第一部分; columnaxes; columnaxis / display=none discreteopts=(colorbands=odd colorbandsattrs=(transparency=0.2)); endcolumnaxes;
*第二部分; layout overlay / yaxisopts=(griddisplay=on offsetmax=0.15 tickvalueattrs=(size=8) labelattrs=(size=7)) walldisplay=none; barchartparm category=j response=duration / datalabel=duration fillattrs=graphdata1 datalabelattrs=(size=7) dataskin=pressed /*displaybaseline=auto*/; scatterplot x=j y=dropped / markerattrs=(symbol=diamondfilled size=11) filledoutlinedmarkers=true markerfillattrs=(color=gold) markeroutlineattrs=(color=black) name='d' legendlabel='Discontinued'; discretelegend 'd' / location=inside valign=top halign=left valueattrs=(size=9) border=false autoitemsize=true; endlayout;
*第三部分; layout overlay / yaxisopts=(griddisplay=on tickvalueattrs=(size=8) labelattrs=(size=7) offsetmax=0 linearopts=(tickvaluepriority=true) label='Change from Baseline (%)') walldisplay=none; bandplot x=j limitupper=20 limitlower=-30 / extend=true display=(outline) outlineattrs=graphdata1(pattern=dash thickness=1); barchartparm category=j response=response / group=drug groupdisplay=cluster datalabelattrs=(size=5) dataskin=pressed name='a' datalabelfitpolicy=rotate; textplot x=j y=codeloc text=code / rotate=90 position=left textattrs=(size=8) contributeoffsets=(ymin); discretelegend 'a' / location=inside valign=bottom halign=left across=2 opaque=false valueattrs=(size=7) border=false; endlayout;
*第四部分; layout overlay / yaxisopts=(griddisplay=on tickvalueattrs=(size=8) labelattrs=(size=7) offsetmax=0 linearopts=(tickvaluepriority=true) reverse=true) walldisplay=none; seriesplot x=j y=baseline / lineattrs=graphdata2(pattern=solid thickness=2); scatterplot x=j y=baseline / markerattrs=(symbol=circlefilled size=7) filledoutlinedmarkers=true markerfillattrs=(color=white) markeroutlineattrs=graphdata2; endlayout; endlayout; endgraph; end; run; ods graphics / reset width=9in height=5in imagename='Waterfall_Baseline_Series'; proc sgrender template=Waterfall_Baseline_Series data=tumorsorted dattrmap=attrmap; format duration 3.0; label baseline='Baseline (mm)'; dattrvar drug="Resp"; run; /*--WaterFall with Tumor Baseline--*/ proc template; define statgraph Waterfall_Baseline_Needle; begingraph / ; entrytitle 'Tumor Response and Duration by Subject Id'; entryfootnote halign=left 'This graph uses simulated data for illustration only' / textattrs=(size=7pt style=italic); layout lattice / columndatarange=union rowweights=(0.3 0.45 0.25) rowgutter=0; columnaxes; columnaxis / display=none discreteopts=(colorbands=odd colorbandsattrs=(transparency=0.2)); endcolumnaxes; layout overlay / yaxisopts=(griddisplay=on offsetmax=0.15 tickvalueattrs=(size=7) labelattrs=(size=7)) walldisplay=none; barchartparm category=j response=duration / datalabel=duration fillattrs=graphdata1 datalabelattrs=(size=5) dataskin=pressed /*displaybaseline=auto*/; scatterplot x=j y=dropped / markerattrs=(symbol=diamondfilled size=9) filledoutlinedmarkers=true markerfillattrs=(color=gold) markeroutlineattrs=(color=black) name='d' legendlabel='Discontinued'; discretelegend 'd' / location=inside valign=top halign=left valueattrs=(size=7) border=false autoitemsize=true; endlayout; layout overlay / yaxisopts=(griddisplay=on tickvalueattrs=(size=5) labelattrs=(size=7) offsetmax=0 linearopts=(tickvaluepriority=true) label='Change from Baseline (%)') walldisplay=none; bandplot x=j limitupper=20 limitlower=-30 / extend=true display=(outline) outlineattrs=graphdata1(pattern=dash thickness=1); barchartparm category=j response=response / group=drug groupdisplay=cluster datalabelattrs=(size=5) dataskin=pressed name='a' datalabelfitpolicy=rotate; textplot x=j y=codeloc text=code / rotate=90 position=left textattrs=(size=6) contributeoffsets=(ymin); discretelegend 'a' / location=inside valign=bottom halign=left across=2 opaque=false valueattrs=(size=5) border=false; endlayout; layout overlay / yaxisopts=(griddisplay=on tickvalueattrs=(size=5) labelattrs=(size=7) offsetmin=0 linearopts=(tickvaluepriority=true)) walldisplay=none; needleplot x=j y=baseline / lineattrs=(color=green pattern=solid thickness=3); scatterplot x=j y=baseline / markerattrs=(symbol=circlefilled size=7) filledoutlinedmarkers=true markerfillattrs=(color=white) markeroutlineattrs=(color=green); endlayout; endlayout; endgraph; end; run; ods graphics / reset width=4in height=4in imagename='Waterfall_Baseline_Needle'; proc sgrender template=Waterfall_Baseline_Needle data=tumorsorted dattrmap=attrmap; format duration 3.0; label baseline='Baseline (mm)'; dattrvar drug="Resp"; run;
本文来自博客园,作者:Iving,转载请注明原文链接:https://www.cnblogs.com/SAS-T/p/15375785.html


 浙公网安备 33010602011771号
浙公网安备 33010602011771号