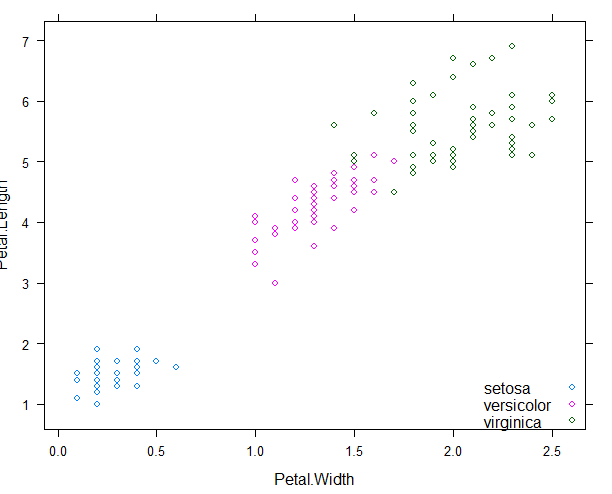
library(lattice)
xyplot(Petal.Length ~ Petal.Width, data = iris, groups = Species,
+ auto.key=list(corner=c(1,0)))
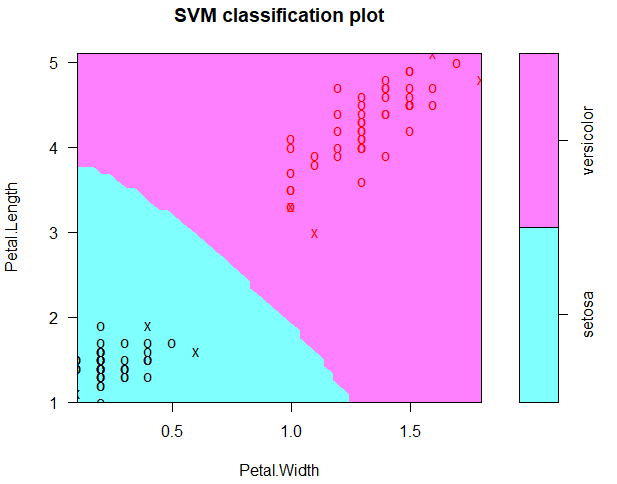
data("iris")
attach(iris)
subdata<-iris[iris$Species != 'virginica',]
subdata$Species<-factor(subdata$Species)
library(e1071)
model1<-svm(Species ~ Petal.Length+Petal.Width,data=subdata)
plot(model1, subdata, Petal.Length ~ Petal.Width)
> model2 <- svm(Species ~ ., data = iris)
> summary(model2)
Call:
svm(formula = Species ~ ., data = iris)
Parameters:
SVM-Type: C-classification
SVM-Kernel: radial
cost: 1
gamma: 0.25
Number of Support Vectors: 51
( 8 22 21 )
Number of Classes: 3
Levels:
setosa versicolor virginica
> x= iris[,5]
> x= iris[,-5]
> y= iris[,5]
> model3 = svm(x,y,kernel="radial",gamma = if(is.vector(x)) 1 else 1/ncol(x))
> pred<-predict(model3,x)
> table(pred,y)
y
pred setosa versicolor virginica
setosa 50 0 0
versicolor 0 48 2
virginica 0 2 48
> pred<-predict(model3,x,decision.values = TRUE)
> attr(pred,"decision.values")[1:4,]
setosa/versicolor setosa/virginica versicolor/virginica
1 1.196152 1.091757 0.6708810
2 1.064621 1.056185 0.8483518
3 1.180842 1.074542 0.6439798
4 1.110699 1.053012 0.6782041
> attr(pred,"decision.values")[77:78,]
setosa/versicolor setosa/virginica versicolor/virginica
77 -1.023056 -0.892961 0.82644637
78 -1.099844 -1.034654 -0.03459913
> pred[77:78]
77 78
versicolor virginica
Levels: setosa versicolor virginica
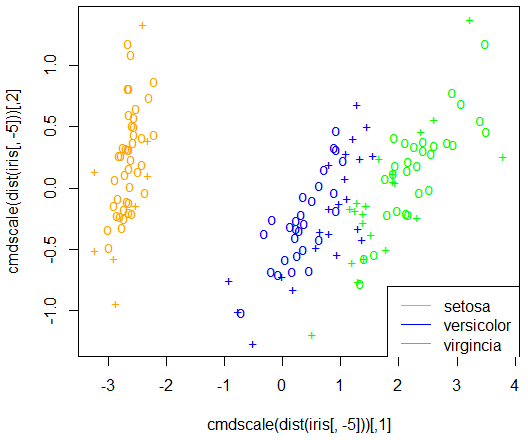
> plot(cmdscale(dist(iris[,-5])),
+ col = c("orange","blue","green")[as.integer(iris[,5])],
+ pch = c("o","+")[1:150 %in% model3$index + 1])
> legend(1.8, -0.8, c("setosa","versicolor","virgincia"),
+ col = c("orange","blue","green"), lty = 1)