时间序列异常检测常见方法和代码
0.介绍
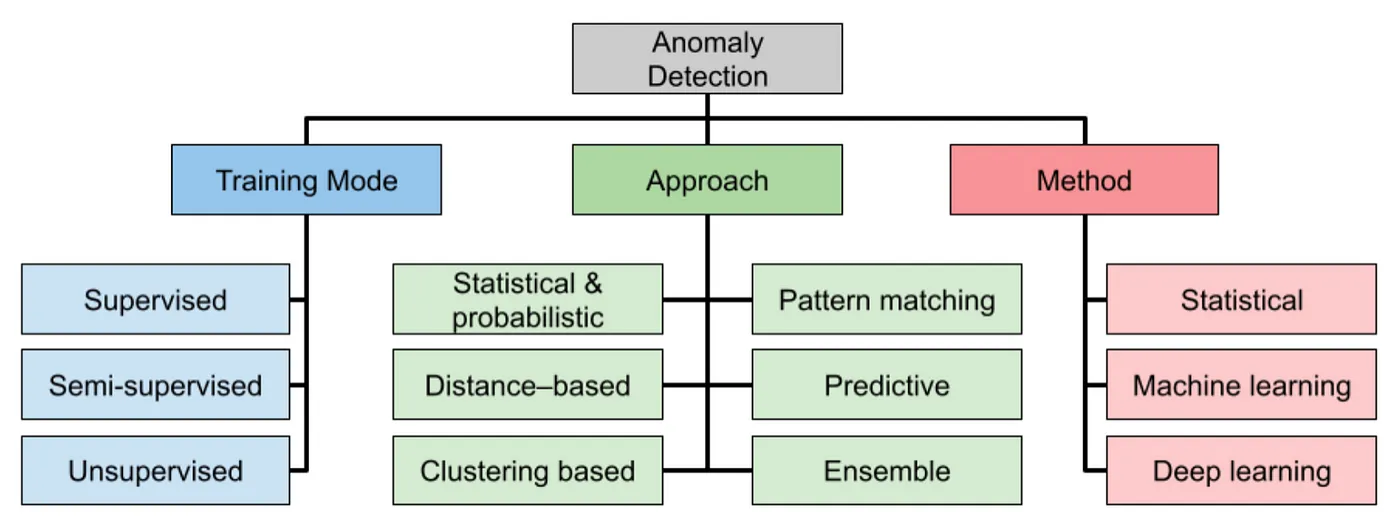
时间序列的异常检测是在序列中识别明显偏离正常情况的数据,从而知晓仪器损坏、欺诈活动、系统故障或其他意外事件。我们介绍监督和无监督学习方法探索各种异常检测技术。
1.了解异常检测
异常通常称为异常值或异常值,是显着偏离数据集中的预期或正常行为的数据点或观察结果。这些偏差可能是由多种因素引起的[1]。
2.用于异常检测的监督学习
监督异常检测模型旨在使用标记数据检测数据集中的异常,其中每个数据点被分类为正常或异常。这些模型通常在带有异常和正常数据点标记示例的数据集上进行训练。以下是一些常用的监督异常检测模型:
2.1隔离森林(Isolation Forest):
- 一种集成方法,通过构建随机森林并隔离需要在树中进行较少分割的数据点来隔离异常。
- 这是一种简单而有效的异常检测方法。
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import IsolationForest
# Generate synthetic data with anomalies
np.random.seed(42)
normal_data = np.random.randn(300, 2)
anomalies = 4 + 1.5 * np.random.randn(10, 2) # Generating anomalies far from normal data
# Combine normal and anomaly data
data = np.vstack([normal_data, anomalies])
# Initialize and fit the Isolation Forest model
clf = IsolationForest(contamination=0.05, random_state=42) # Adjust contamination parameter as needed
clf.fit(data)
# Predict whether each data point is an anomaly (1 for inliers, -1 for outliers)
predictions = clf.predict(data)
# Visualize the results
plt.scatter(data[:, 0], data[:, 1], c=predictions, cmap='viridis')
plt.colorbar(label="Anomaly Score")
plt.title("Anomaly Detection using Isolation Forest")
plt.show()
# Identify anomalies (outliers)
anomalies_indices = np.where(predictions == -1)[0]
print("Detected anomalies:", anomalies_indices)
2.2k-最近邻 (KNN):
- 通过使用到第 k 个最近邻的距离作为异常的度量,KNN 可以适用于异常检测。
- 如果数据点与其 k 最近邻显着不同,则将数据点分类为异常。
import numpy as np
import matplotlib.pyplot as plt
from sklearn.neighbors import KNeighborsClassifier
# Generate synthetic data with anomalies
np.random.seed(42)
normal_data = np.random.randn(300, 2)
anomalies = 4 + 1.5 * np.random.randn(10, 2) # Generating anomalies far from normal data
# Combine normal and anomaly data
data = np.vstack([normal_data, anomalies])
# Label the data (1 for normal data, 0 for anomalies)
labels = np.concatenate([np.ones(len(normal_data)), np.zeros(len(anomalies))])
# Initialize and fit the KNN model
k = 5 # Adjust the number of neighbors as needed
clf = KNeighborsClassifier(n_neighbors=k)
clf.fit(data, labels)
# Predict whether each data point is an anomaly (0 for anomalies, 1 for normal data)
predictions = clf.predict(data)
# Visualize the results
plt.scatter(data[:, 0], data[:, 1], c=predictions, cmap='viridis')
plt.colorbar(label="Anomaly Score")
plt.title("Anomaly Detection using KNN")
plt.show()
# Identify anomalies (outliers)
anomalies_indices = np.where(predictions == 0)[0]
print("Detected anomalies:", anomalies_indices)
2.3SVM(支持向量机):
- 单类 SVM 是一种监督算法,可以学习区分多数类(正常)和少数类(异常)。
- 它构建了一个超平面,将正常数据点与潜在异常值分开。
import numpy as np
import matplotlib.pyplot as plt
from sklearn.svm import OneClassSVM
# Generate synthetic data with anomalies
np.random.seed(42)
normal_data = np.random.randn(300, 2)
anomalies = 4 + 1.5 * np.random.randn(10, 2) # Generating anomalies far from normal data
# Combine normal and anomaly data
data = np.vstack([normal_data, anomalies])
# Initialize and fit the One-Class SVM model
clf = OneClassSVM(nu=0.05, kernel="rbf") # Adjust the nu parameter as needed
clf.fit(data)
# Predict whether each data point is an anomaly (-1 for anomalies, 1 for normal data)
predictions = clf.predict(data)
# Visualize the results
plt.scatter(data[:, 0], data[:, 1], c=predictions, cmap='viridis')
plt.colorbar(label="Anomaly Score")
plt.title("Anomaly Detection using One-Class SVM")
plt.show()
# Identify anomalies (outliers)
anomalies_indices = np.where(predictions == -1)[0]
print("Detected anomalies:", anomalies_indices)
2.4随机森林:
- 虽然随机森林通常用于分类任务,但它们也可以用于监督异常检测,将一类视为异常数据,将另一类视为正常数据。
2.5集成方法:
- AdaBoost 和 Gradient Boosting 等集成方法可通过组合多个弱学习器来识别异常,从而用于异常检测。
2.6决策树:
- 通过训练树根据数据点的特征将数据点分类为正常或异常,决策树可以适用于监督异常检测。
2.7极值理论(EVT):
- EVT 模型用于对数据的尾部分布进行建模,并通过将极值与建模分布进行比较来检测极值的异常。
2.8支持向量数据描述(SVDD):
- SVDD 是支持向量机的一种变体,它围绕正常数据点构建超球面,并将异常分类为超球面之外的点。
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import OneClassSVM
from sklearn.ensemble import IsolationForest
from sklearn.datasets import make_classification
from scipy.stats import norm
# Generate synthetic data with anomalies
np.random.seed(42)
normal_data, _ = make_classification(n_samples=300, n_features=2, n_informative=2, n_redundant=0, random_state=42)
anomalies = np.random.randn(10, 2) * 3 + np.array([4, 4]) # Generating anomalies far from normal data
data = np.vstack([normal_data, anomalies])
# Labels (1 for normal data, -1 for anomalies)
labels = np.array([1] * len(normal_data) + [-1] * len(anomalies))
# Initialize and fit Random Forest classifier for anomaly detection
rf = RandomForestClassifier(n_estimators=100, random_state=42)
rf.fit(data, labels)
rf_predictions = rf.predict(data)
# Initialize and fit AdaBoost classifier for anomaly detection
ada = AdaBoostClassifier(n_estimators=100, random_state=42)
ada.fit(data, labels)
ada_predictions = ada.predict(data)
# Initialize and fit Gradient Boosting classifier for anomaly detection
gb = GradientBoostingClassifier(n_estimators=100, random_state=42)
gb.fit(data, labels)
gb_predictions = gb.predict(data)
# Initialize and fit Decision Tree classifier for anomaly detection
dt = DecisionTreeClassifier(random_state=42)
dt.fit(data, labels)
dt_predictions = dt.predict(data)
# Initialize and fit One-Class SVM for anomaly detection
svm = OneClassSVM(kernel='rbf', nu=0.05)
svm.fit(data)
svm_predictions = svm.predict(data)
# Initialize and fit Isolation Forest for anomaly detection
iso_forest = IsolationForest(contamination=0.05, random_state=42)
iso_forest.fit(data)
iso_forest_predictions = iso_forest.predict(data)
# Initialize and fit Extreme Value Theory (EVT) for anomaly detection
mu, sigma = norm.fit(data) # Estimate distribution parameters
threshold = np.percentile(norm.pdf(data, mu, sigma), 95) # Set a threshold
evt_predictions = (norm.pdf(data, mu, sigma) < threshold).astype(int)
# Visualize the results
def plot_anomaly_detection_results(predictions, title):
plt.scatter(data[:, 0], data[:, 1], c=predictions, cmap='viridis')
plt.colorbar(label="Anomaly Score")
plt.title(title)
plt.show()
plot_anomaly_detection_results(rf_predictions, "Anomaly Detection using Random Forest")
plot_anomaly_detection_results(ada_predictions, "Anomaly Detection using AdaBoost")
plot_anomaly_detection_results(gb_predictions, "Anomaly Detection using Gradient Boosting")
plot_anomaly_detection_results(dt_predictions, "Anomaly Detection using Decision Trees")
plot_anomaly_detection_results(svm_predictions, "Anomaly Detection using One-Class SVM")
plot_anomaly_detection_results(iso_forest_predictions, "Anomaly Detection using Isolation Forest")
2.9XGBoost:
- XGBoost 是梯度增强的扩展,可以将一类视为异常数据,将另一类视为正常数据,用作监督异常检测算法。
2.10神经网络:
- 深度学习技术,例如前馈神经网络和循环神经网络(RNN),可以被训练为监督异常检测器。
- 自动编码器是一种神经网络,通常用于无监督的异常检测,但也可以以监督的方式进行训练。
2.11逻辑回归:
- 通过学习分离正常数据点和异常数据点的决策边界,逻辑回归可用于监督异常检测。
2.12朴素贝叶斯:
- 朴素贝叶斯分类器可用于通过对正常数据的概率分布进行建模并识别与其偏差来进行监督异常检测。
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from sklearn.ensemble import IsolationForest
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import confusion_matrix, classification_report
from xgboost import XGBClassifier
from keras.models import Sequential
from keras.layers import Dense
# Generate synthetic data with anomalies
np.random.seed(42)
X, y = make_classification(n_samples=500, n_features=2, n_classes=2, n_clusters_per_class=1, n_redundant=0)
anomalies = 2 + 0.5 * np.random.randn(10, 2) # Generating anomalies far from normal data
X = np.vstack([X, anomalies])
y = np.hstack([y, np.ones(len(anomalies))]) # Anomalies are labeled as 1, normal data as 0
# Split the data into train and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Isolation Forest for Anomaly Detection
iforest = IsolationForest(contamination=0.05, random_state=42)
iforest.fit(X_train)
y_iforest_pred = iforest.predict(X_test)
# Logistic Regression for Anomaly Detection
lr = LogisticRegression()
lr.fit(X_train, y_train)
y_lr_pred = lr.predict(X_test)
# Naive Bayes for Anomaly Detection
nb = GaussianNB()
nb.fit(X_train, y_train)
y_nb_pred = nb.predict(X_test)
# XGBoost for Anomaly Detection
xgb = XGBClassifier()
xgb.fit(X_train, y_train)
y_xgb_pred = xgb.predict(X_test)
# Neural Network for Anomaly Detection
nn_model = Sequential()
nn_model.add(Dense(8, input_dim=2, activation='relu'))
nn_model.add(Dense(1, activation='sigmoid'))
nn_model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
nn_model.fit(X_train, y_train, epochs=50, batch_size=10, verbose=0)
y_nn_pred = nn_model.predict(X_test)
y_nn_pred = (y_nn_pred > 0.5).astype(int)
# Evaluation
models = ["Isolation Forest", "Logistic Regression", "Naive Bayes", "XGBoost", "Neural Network"]
predictions = [y_iforest_pred, y_lr_pred, y_nb_pred, y_xgb_pred, y_nn_pred]
for model, y_pred in zip(models, predictions):
print(f"Model: {model}")
print("Confusion Matrix:")
print(confusion_matrix(y_test, y_pred))
print("Classification Report:")
print(classification_report(y_test, y_pred))
print("="*50)
# Visualization of Results (for Neural Network)
plt.scatter(X_test[y_test == 0][:, 0], X_test[y_test == 0][:, 1], c='b', label='Normal Data')
plt.scatter(X_test[y_test == 1][:, 0], X_test[y_test == 1][:, 1], c='r', label='Anomalies')
plt.title('Neural Network Anomaly Detection')
plt.legend()
plt.show()
3.无监督学习技术
3.1基于密度的方法
- DBSCAN 在识别高密度区域中的数据点簇,同时将低密度区域中的数据点标记为异常或噪声方面特别有效。它基于数据密度的概念进行操作,使其对不规则形状的集群具有鲁棒性,并且能够处理具有不同集群大小的数据集。以下是 DBSCAN 的工作原理以及使用 scikit-learn 的示例:
from sklearn.cluster import DBSCAN
from sklearn.datasets import make_moons
import matplotlib.pyplot as plt
import numpy as np
# Create a synthetic dataset with two moon-shaped clusters
X, _ = make_moons(n_samples=300, noise=0.6, random_state=42)
# Initialize and fit DBSCAN
dbscan = DBSCAN(eps=0.3, min_samples=5)
dbscan.fit(X)
# Get cluster labels (-1 indicates anomalies)
labels = dbscan.labels_
# Separate data points into clusters and anomalies
core_samples_mask = np.zeros_like(labels, dtype=bool)
core_samples_mask[dbscan.core_sample_indices_] = True
n_clusters_ = len(set(labels)) - (1 if -1 in labels else 0)
n_noise_ = list(labels).count(-1)
# Plot the clusters and anomalies
unique_labels = set(labels)
colors = [plt.cm.Spectral(each) for each in np.linspace(0, 1, len(unique_labels))]
for k, col in zip(unique_labels, colors):
if k == -1:
col = [0, 0, 0, 1] # Black color for anomalies
class_member_mask = (labels == k)
xy = X[class_member_mask & core_samples_mask]
plt.plot(xy[:, 0], xy[:, 1], 'o', markerfacecolor=tuple(col), markersize=6)
xy = X[class_member_mask & ~core_samples_mask]
plt.plot(xy[:, 0], xy[:, 1], 'o', markerfacecolor=tuple(col), markersize=6)
plt.title(f'Estimated number of clusters: {n_clusters_}\nEstimated number of anomalies: {n_noise_}')
plt.show()
3.2高斯混合模型 (GMM)
- GMM 广泛用于聚类和密度估计任务,但它们也可以通过识别建模分布下可能性较低的数据点来应用于异常检测。以下是 GMM 如何进行异常检测的说明以及使用 scikit-learn 的
GaussianMixture的代码示例。
import numpy as np
from sklearn.mixture import GaussianMixture
import matplotlib.pyplot as plt
# Generate a synthetic dataset with anomalies
np.random.seed(42)
normal_data = np.random.randn(300, 2)
anomalies = 4 + 1.5 * np.random.randn(10, 2) # Generating anomalies far from normal data
# Combine normal and anomaly data
data = np.vstack([normal_data, anomalies])
# Train a Gaussian Mixture Model (GMM)
n_components = 5 # Number of Gaussian components
gmm = GaussianMixture(n_components=n_components, covariance_type='full', random_state=42)
gmm.fit(data)
# Calculate the likelihood scores for each data point
likelihoods = -gmm.score_samples(data)
# Set a threshold for anomaly detection (e.g., using a percentile)
threshold = np.percentile(likelihoods, 95) # Adjust the percentile as needed
# Identify anomalies based on the threshold
anomalies_indices = np.where(likelihoods < threshold)[0]
# Plot the data and highlight anomalies
plt.scatter(data[:, 0], data[:, 1], c='b', label='Normal Data')
plt.scatter(data[anomalies_indices, 0], data[anomalies_indices, 1], c='r', marker='x', label='Anomalies')
plt.legend()
plt.title('Anomaly Detection using Gaussian Mixture Model')
plt.show()
3.3自动编码器
自动编码器由编码器和解码器网络组成,其主要目的是学习输入数据的紧凑表示。可以通过比较自动编码器的重构误差(输入和输出之间的差异)来检测异常。当模型遇到与学习模式显着偏差的数据时,重建误差往往很高,表明存在异常。以下介绍了用于无监督异常检测的自动编码器,以及使用 TensorFlow 和 PyTorch 的代码示例。
import numpy as np
import tensorflow as tf
from tensorflow import keras
# Generate a synthetic dataset with anomalies
np.random.seed(42)
normal_data = np.random.randn(300, 10)
anomalies = 4 + 1.5 * np.random.randn(10, 10) # Generating anomalies far from normal data
# Combine normal and anomaly data
data = np.vstack([normal_data, anomalies])
# Build an autoencoder model
input_dim = data.shape[1]
encoding_dim = 5 # Adjust the bottleneck layer size as needed
model = keras.Sequential([
keras.layers.Input(shape=(input_dim,)),
keras.layers.Dense(encoding_dim, activation='relu'),
keras.layers.Dense(input_dim, activation='linear') # Linear activation for reconstruction
])
model.compile(optimizer='adam', loss='mse')
# Train the autoencoder
model.fit(data, data, epochs=100, batch_size=32, shuffle=True)
# Calculate reconstruction errors
reconstructed_data = model.predict(data)
reconstruction_errors = np.mean(np.square(data - reconstructed_data), axis=1)
# Set a threshold for anomaly detection (e.g., using a percentile)
threshold = np.percentile(reconstruction_errors, 95) # Adjust the percentile as needed
# Identify anomalies based on the threshold
anomalies_indices = np.where(reconstruction_errors > threshold)[0]
print("Detected anomalies:", anomalies_indices)
以下是使用 PyTorch 训练自动编码器和检测异常的代码示例:
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
# Generate a synthetic dataset with anomalies
np.random.seed(42)
normal_data = np.random.randn(300, 10)
anomalies = 4 + 1.5 * np.random.randn(10, 10) # Generating anomalies far from normal data
# Combine normal and anomaly data
data = np.vstack([normal_data, anomalies])
# Define an autoencoder class
class Autoencoder(nn.Module):
def __init__(self, input_dim, encoding_dim):
super(Autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Linear(input_dim, encoding_dim),
nn.ReLU()
)
self.decoder = nn.Sequential(
nn.Linear(encoding_dim, input_dim)
)
def forward(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
# Create an instance of the autoencoder
input_dim = data.shape[1]
encoding_dim = 5 # Adjust the bottleneck layer size as needed
autoencoder = Autoencoder(input_dim, encoding_dim)
# Define loss function and optimizer
criterion = nn.MSELoss()
optimizer = optim.Adam(autoencoder.parameters(), lr=0.001)
# Convert data to PyTorch tensors
data_tensor = torch.Tensor(data)
# Train the autoencoder
num_epochs = 100
for epoch in range(num_epochs):
optimizer.zero_grad()
outputs = autoencoder(data_tensor)
loss = criterion(outputs, data_tensor)
loss.backward()
optimizer.step()
# Calculate reconstruction errors
reconstructed_data = autoencoder(data_tensor).detach().numpy()
reconstruction_errors = np.mean(np.square(data - reconstructed_data), axis=1)
# Set a threshold for anomaly detection (e.g., using a percentile)
threshold = np.percentile(reconstruction_errors, 95) # Adjust the percentile as needed
# Identify anomalies based on the threshold
anomalies_indices = np.where(reconstruction_errors > threshold)[0]
print("Detected anomalies:", anomalies_indices)
3.2聚类的算法:
- 谱聚类是一种基于图的方法,可通过将与主聚类连接性较低的数据点视为异常来用于异常检测。
3.2.1SVM(支持向量机):
-
One-Class SVM 构建了一个超平面,将大部分数据与潜在异常区分开来。
— 当异常罕见且分布不均匀时,它特别有用。
3.2.2局部离群因子 (LOF):
- LOF 计算数据点相对于其邻居的密度。密度明显低于其邻居的数据点被视为异常。
- 它可以有效识别数据集中的局部异常。
3.2.3隔离内核(iKernel):
- iKernel 是隔离森林的扩展,它使用核方法来捕获数据中的非线性关系。
- 它适用于具有复杂非线性结构的数据集。
3.2.4椭圆形信封:
- 椭圆包络将多元高斯分布拟合到数据,并根据与估计分布的马哈拉诺比斯距离识别异常。
- 它对于检测多元异常值特别有用。
3.2.5基于直方图的异常值检测 (HBOS):
- HBOS将特征空间离散成直方图,并根据直方图的稀疏性计算异常分数。
- 它非常高效并且适用于高维数据。
3.2.6基于聚类的方法:
- 各种聚类技术(例如层次聚类或 k 均值聚类)可用于通过将小聚类中的数据点视为异常来进行异常检测。
在运行以下脚本之前,请确保安装 pyod 库。
pip install pyod
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_blobs
from sklearn.cluster import SpectralClustering, KMeans
from sklearn.svm import OneClassSVM
from sklearn.neighbors import LocalOutlierFactor
from sklearn.ensemble import IsolationForest
from sklearn.covariance import EllipticEnvelope
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import pairwise_distances_argmin_min
from pyod.models.hbos import HBOS
# Generate synthetic data with anomalies
np.random.seed(42)
n_samples = 300
n_features = 2
n_outliers = 10
X, _ = make_blobs(n_samples=n_samples, n_features=n_features, centers=1, random_state=42)
X[:n_outliers] = 10 + 2 * np.random.randn(n_outliers, n_features) # Adding anomalies
# Standardize the data
scaler = StandardScaler()
X = scaler.fit_transform(X)
# Create subplots for each method
fig, axs = plt.subplots(2, 3, figsize=(15, 10))
# Spectral Clustering
spectral = SpectralClustering(n_clusters=1, affinity="nearest_neighbors")
spectral.fit(X)
labels_spectral = spectral.labels_
axs[0, 0].scatter(X[:, 0], X[:, 1], c=labels_spectral, cmap='viridis')
axs[0, 0].set_title("Spectral Clustering")
# One-Class SVM
ocsvm = OneClassSVM(nu=0.05, kernel="rbf")
ocsvm.fit(X)
labels_ocsvm = ocsvm.predict(X)
axs[0, 1].scatter(X[:, 0], X[:, 1], c=labels_ocsvm, cmap='viridis')
axs[0, 1].set_title("One-Class SVM")
# Local Outlier Factor (LOF)
lof = LocalOutlierFactor(n_neighbors=20, contamination=0.05)
labels_lof = lof.fit_predict(X)
axs[0, 2].scatter(X[:, 0], X[:, 1], c=labels_lof, cmap='viridis')
axs[0, 2].set_title("Local Outlier Factor")
# Isolation Kernel
kernel = KMeans(n_clusters=1, random_state=42)
kernel.fit(X)
labels_kernel, _ = pairwise_distances_argmin_min(X, kernel.cluster_centers_)
axs[1, 0].scatter(X[:, 0], X[:, 1], c=labels_kernel, cmap='viridis')
axs[1, 0].set_title("Isolation Kernel")
# Elliptic Envelope
elliptic = EllipticEnvelope(contamination=0.05)
labels_elliptic = elliptic.fit_predict(X)
axs[1, 1].scatter(X[:, 0], X[:, 1], c=labels_elliptic, cmap='viridis')
axs[1, 1].set_title("Elliptic Envelope")
# HBOS (Histogram-Based Outlier Detection)
hbos = HBOS(contamination=0.05)
labels_hbos = hbos.fit_predict(X)
axs[1, 2].scatter(X[:, 0], X[:, 1], c=labels_hbos, cmap='viridis')
axs[1, 2].set_title("HBOS")
plt.show()
无监督异常检测算法的选择取决于数据的特征、所需的可解释性水平以及可用的计算资源。通常建议尝试多种算法并评估它们在特定数据集上的性能,以选择最合适的方法。
4.结论
我们简要涵盖了异常检测的各个方面,从基础的概念到使用 scikit-learn 和其他库实现监督和无监督学习技术。
W. Fang, Y. Shao, P.E.D. Love, T. Hartmann, W. Liu, Detecting anomalies and de-noising monitoring data from sensors: A smart data approach, Advanced Engineering Informatics 55 (2023) 101870. https://doi.org/10.1016/j.aei.2022.101870. ↩︎
-------------------------------------------
个性签名:独学而无友,则孤陋而寡闻。做一个灵魂有趣的人!
如果觉得这篇文章对你有小小的帮助的话,记得在右下角点个“推荐”哦,博主在此感谢!
万水千山总是情,打赏一分行不行,所以如果你心情还比较高兴,也是可以扫码打赏博主,哈哈哈(っ•̀ω•́)っ✎⁾⁾!


 时间序列的异常检测是在序列中识别明显偏离正常情况的数据,从而知晓仪器损坏、欺诈活动、系统故障或其他意外事件。我们介绍监督和无监督学习方法探索各种异常检测技术。
时间序列的异常检测是在序列中识别明显偏离正常情况的数据,从而知晓仪器损坏、欺诈活动、系统故障或其他意外事件。我们介绍监督和无监督学习方法探索各种异常检测技术。

