20180711-统计PDB中的蛋白质种类、膜蛋白文件个数及信息等
20180710完成这份工作。简单,但是完成了还是很开心。在我尝试如何使用pickle保存数据后,尝试保存PDB文件中“HEADER”中的信息。文件均保存于实验室服务器(97.73.198.168)/home/RaidDisk/BiodataTest/zyh_pdb_test/tests路径下。本文将记录流程并分享统计结果。
先插入一段代码看看
from PDBParseBase import PDBParserBase import os import json import datetime from DBParser import ParserBase import pickle def length_counter(seqres_info): pdb_id = seqres_info['pdb_id'] number = 0 for item in seqres_info.keys(): if item != 'pdb_id' and item != 'SEQRES_serNum': number += int(seqres_info[item]['SEQRES_numRes']) pass #print(pdb_id) #print(number) id_and_lenth = [] id_and_lenth.append(pdb_id) id_and_lenth.append(number) return id_and_lenth def find_all_headers(rootdir,saveFilePath,saveFilePath2): #with the help of PDBParserBase,just put HEADER inf into a pickle with the form of list pdbbase = PDBParserBase() start = datetime.datetime.now() count = 0 f = open(saveFilePath,'wb') f2 = open(saveFilePath2,'wb') header_info_all = [] for parent,dirnames,filenames in os.walk(rootdir): #print(dirnames) #print(filenames) for filename in filenames: count = count + 1 try: PDBfile = filename #print(filename) header_info = pdbbase.get_header_info(os.path.join(parent,filename)) #print(header_info) header_info_all.append(header_info) if count %1000== 0: print(count) end = datetime.datetime.now() print (end-start) pass except: print(filename) end = datetime.datetime.now() print (end-start) pickle.dump(filename, f2) #pdb1ov7.ent else: if count %1111 == 0: print(count) pickle.dump(header_info_all, f,protocol=2) end = datetime.datetime.now() print (end-start) print("Done") return header_info_all def hd_ctr_clsfcsh(listpath): #header_counter_classfication #return a dic ,the key is the classfication and the value is the num with open(listpath, 'rb') as f: header_list = pickle.load(f) #print(header_list) dic = {} for item in header_list: classification = item['HEADER_classification'] print (classification) if 'MEMBRAN' in classification: if classification in dic.keys(): dic[classification] = dic[classification] +1 else: dic[classification] = 1 else: pass dict= sorted(dic.items(), key=lambda d:d[1], reverse = True) print(dict) return dict def get_filenames(listpath,keyword,resultpath): with open(listpath, 'rb') as f: header_list = pickle.load(f) filenames = [] for item in header_list: classification = item['HEADER_classification'] if keyword in classification: #print(item) #print (item['pdb_id']) #print (classification) id = item['pdb_id'] filenames.append(id) else: #print ('dad') pass print(len(filenames)) print(filenames) f2 = open(resultpath,'wb') pickle.dump(filenames, f2,protocol=2) print("done") return filenames if __name__ == "__main__": rootdir = "/home/BiodataTest/updb" #1 is to save list 2 is to save some wrang message saveFilePath = "/home/BiodataTest/test_picale/header_counter.pic" saveFilePath2 = "/home/BiodataTest/test_picale/header_counter_wrang.pic" #all_header_list = find_all_headers(rootdir,saveFilePath,saveFilePath2) HEADER_LIST_FILE = "/home/BiodataTest/test_picale/header_counter_list.pic" hd_ctr_clsfcsh_dic = hd_ctr_clsfcsh(HEADER_LIST_FILE) resultpath = "/home/BiodataTest/test_picale/Membrane_Filename_list.pic" #wanted_filenames = get_filenames(HEADER_LIST_FILE,'MEMBRANE',resultpath)
昨天写的代码,第二天就忘记了。
函数一:find_all_headers(rootdir,saveFilePath,saveFilePath2),输入文件路径,找到PDB文件中所有的“HEADER”信息,其中包括,文件日期、蛋白质种类、蛋白质名字。存入pickle文件中,保存成一个list。难点在于pickle的第三个参数的值是“2”。
函数二:hd_ctr_clsfcsh(listpath):#header_counter_classfication,将刚才输入的list输入,统计出每个分类的数量。输出的是一个字典,键是分类名字,值是该分类的数量。
函数三:get_filenames(listpath,keyword,resultpath),根据关键词获取给定文件的文件名。在这里,我将所有分类这个信息中含有“MEMBRANE”的蛋白质名字找到,保存成列表输出到文件中。(蛋白质名字就是文件名字)
枯燥的代码介绍完了,来点好看的:大饼图。
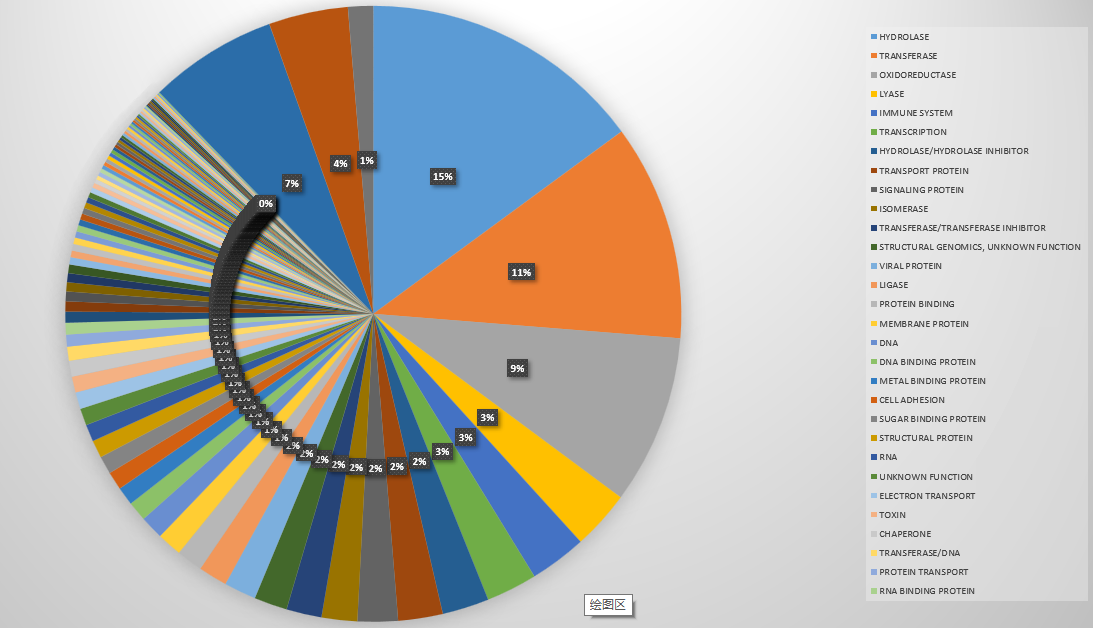
这个饼状图画出了PDB数据库中蛋白质种类的分布,其实是不准确的,比如有的蛋白分别属于膜蛋白和转运蛋白。但是标注为“MEMBRANE PROTEIN, TRANSPORT PROTEIN”,那我们把它归为一类。
我们统计了140946个文件,总共有96(100>x)+400(100>x>9)+1606(10>x>1) +1863(x = 1) = 2965个种类。我们选取前1%看看它们分别是什么:
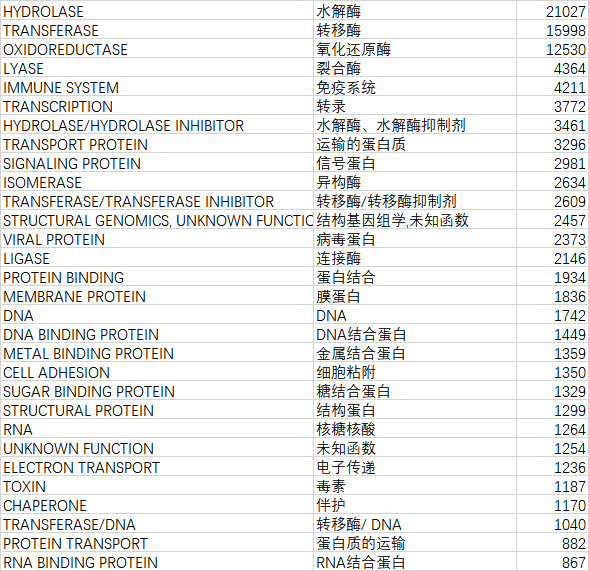
就是它们。总共105057,占总量140946的74.53%。也就是前百分之一的种类占了百分之七十五的数据量。(看起来好残酷的样子哦)
第二部分是有关自己的课题,膜蛋白究竟有多少呢?额,1836个,凄惨的很。所以我扩大了搜索范围,只要包含了“MEMBRANE”的就当作数据提取出来了。注意这个单词的末尾有个“E”,没有"E"的话,数量会增加5个。因为有五个文件叫做“MEMBRANCE PROTEIN”.

找到了它们之后,我们总共获得2335个膜蛋白文件名字,之后将它们解压好,放在预期的文件夹里去。
from PDBParseBase import PDBParserBase import os import json import datetime from DBParser import ParserBase #import DataStorage as DS import time, os,datetime,logging,gzip,configparser,pickle def UnzipFile(fileName,unzipFileName): """""" try: zipHandle = gzip.GzipFile(mode='rb',fileobj=open(fileName,'rb')) with open(unzipFileName, 'wb') as fileHandle: fileHandle.write(zipHandle.read()) except IOError: raise("Unzip file failed!") def mkdir(path): #Created uncompress path folder isExists=os.path.exists(path) if not isExists: os.makedirs(path) print(path + " Created folder sucessful!") return True else: #print ("this path is exist") return False if __name__ == "__main__": #rootdir = "/home/BiodataTest/updb" #rootdir = "/home/BiodataTest/pdb" rootdir = "/home/BiodataTest/membraneprotein" count = 0 countcon = 0 start = datetime.datetime.now() saveFilePath = "/home/BiodataTest/picale_zyh_1000/picale.pic" #Storage = DS.DataStorage('PDB_20180410') mempt_path = "/home/BiodataTest/test_picale/Membrane_Filename_list.pic" with open(mempt_path, 'rb') as f: memprtn_file = pickle.load(f) print(memprtn_file) count = 0 for parent,dirnames,filenames in os.walk(rootdir): for filename in filenames: count = count + 1 #start unzip,get the target name and make a files dirname = filename[4:6] filename_for_membrane_1 = filename[3:7] filename_for_membrane = filename_for_membrane_1.upper() print(filename_for_membrane) if(filename_for_membrane in memprtn_file): print(filename_for_membrane) filename_with_rootdir = "/home/BiodataTest/pdb/pdb/" + str(dirname)+"/"+str(filename) unzipFileName = "/home/BiodataTest/membraneprotein/" + str(dirname)+"/"+str(filename) mkdir("/home/BiodataTest/membraneprotein/" + str(dirname)+"/") try: UnzipFile(filename_with_rootdir,unzipFileName[:-3]) pass except: continue else: #print(filename) pass pass #print(filename) end = datetime.datetime.now() print("alltime = ") print (end-start) print(count) print("Done")
于是就获得了这些膜蛋白。
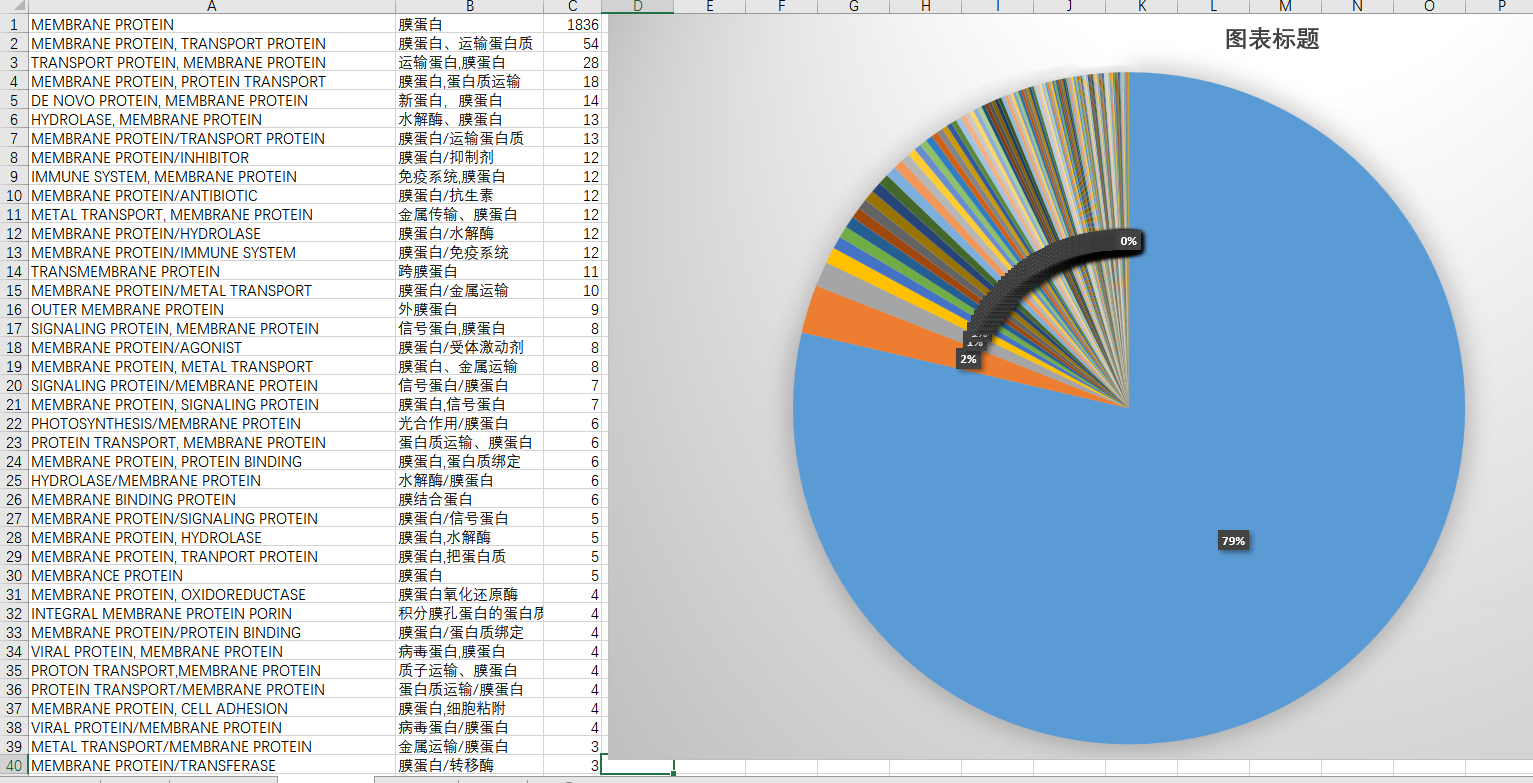
第三十行“MEMBRANCE”的多字母变体英文就大剌剌的显示在那里。。。总结工作也是需要做好的,不然可能之后会忘记吧。


 浙公网安备 33010602011771号
浙公网安备 33010602011771号