LDA与QDA
作者:桂。
时间:2017-05-23 06:37:31
链接:http://www.cnblogs.com/xingshansi/p/6892317.html
前言
仍然是python库函数scikit-learn的学习笔记,内容Regression-1.2Linear and Quadratic Discriminant Analysis部分,主要包括:
1)线性分类判别(Linear discriminant analysis, LDA)
2)二次分类判别(Quadratic discriminant analysis, QDA)
3)Fisher判据
一、线性分类判别
对于二分类问题,LDA针对的是:数据服从高斯分布,且均值不同,方差相同。
概率密度:
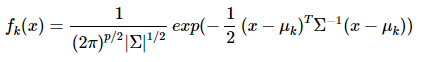
p是数据的维度。
分类判别函数:
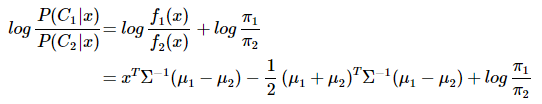
可以看出结果是关于x的一次函数:wx+w0,线性分类判别的说法由此得来。
参数计算:
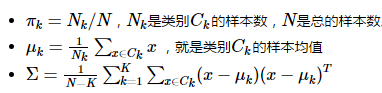
二、二次分类判别
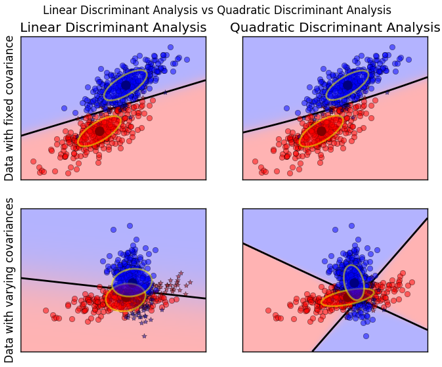
对于二分类问题,QDA针对的是:数据服从高斯分布,且均值不同,方差不同。
数据方差相同的时候,一次判别就可以,如左图所示;但如果方差差别较大,就是一个二次问题了,像右图那样。

从sklearn给的例子中,也容易观察到:

QDA对数据有更好的适用性,QDA判别公式:
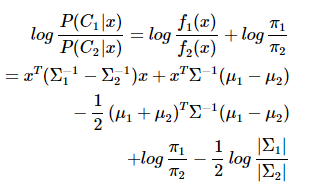
三、Fisher判据
A-Fisher理论推导
Fisher一个总原则是:投影之后的数据,最小化类内误差,同时最大化类间误差
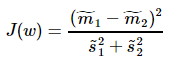
其中, ,
,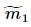 、
、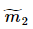 分别对应投影后的类均值。
分别对应投影后的类均值。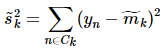 对应投影后的类内方差。
对应投影后的类内方差。
重写类内总方差、类间距离:
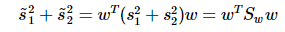
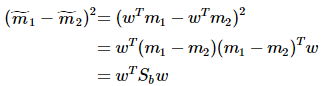
准则函数重写:
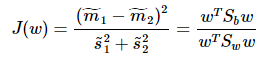
这就是泛化瑞利熵的形式了,容易求解:
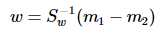
其中 常借助SVD求解:Sw = U∑VT,Sw-1 = U∑-1VT,借助特征值分解也是可以的。
常借助SVD求解:Sw = U∑VT,Sw-1 = U∑-1VT,借助特征值分解也是可以的。
B-LDA与Fisher
这里的LDA指代上面提到的利用概率求解的贝叶斯最优估计器。可以证明:
二分类任务中两类数据满足高斯分布且方差相同时,线性判别分析(指Fisher方法)产生贝叶斯最优分类器(指本文的LDA)。
对于Fisher,求J的最大值:
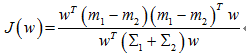
贝叶斯最优,即距离分类中心距离除以两个类中心距离的比值最小:

二者倒数关系,一个最大化,一个最小化,所以是等价的。
补充一句:误差为高斯分布,与最小二乘也是等价的。
这样一来,求解就有了三个思路:奇异值分解SVD,最小二乘Lsqr,特征值分解eigen,这也是Sklearn的思路:

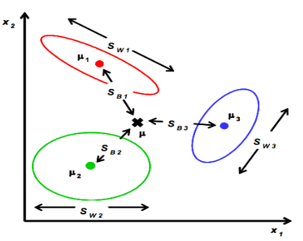
C-多分类LDA
定义类内散度矩阵:
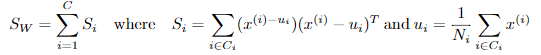
定义类间散度矩阵:
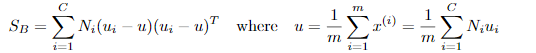
定义总散度矩阵:
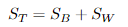
可见ST 、SB、 SW三者任意取两个即可。
准则函数:
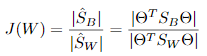
其中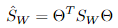 ,
,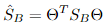 。
。
最优化求解:
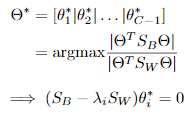
将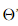 看作投影矩阵,其闭式解是
看作投影矩阵,其闭式解是 的d'个最大非零广义特征值对应的特征向量组成的矩阵。d'通常远小于原数据属性数d,这就实现了监督的降维。
的d'个最大非零广义特征值对应的特征向量组成的矩阵。d'通常远小于原数据属性数d,这就实现了监督的降维。
参数求解利用train data:
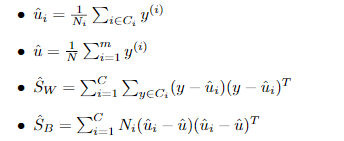
四、Sk-learn基本用法
LDA:
lda = LinearDiscriminantAnalysis(solver="svd", store_covariance=True) y_pred = lda.fit(X, y).predict(X)
QDA:
qda = QuadraticDiscriminantAnalysis(store_covariances=True) y_pred = qda.fit(X, y).predict(X)
LDA与QDA应用实例:
"""
====================================================================
Linear and Quadratic Discriminant Analysis with confidence ellipsoid
====================================================================
Plot the confidence ellipsoids of each class and decision boundary
"""
print(__doc__)
from scipy import linalg
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
from matplotlib import colors
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
###############################################################################
# colormap
cmap = colors.LinearSegmentedColormap(
'red_blue_classes',
{'red': [(0, 1, 1), (1, 0.7, 0.7)],
'green': [(0, 0.7, 0.7), (1, 0.7, 0.7)],
'blue': [(0, 0.7, 0.7), (1, 1, 1)]})
plt.cm.register_cmap(cmap=cmap)
###############################################################################
# generate datasets
def dataset_fixed_cov():
'''Generate 2 Gaussians samples with the same covariance matrix'''
n, dim = 300, 2
np.random.seed(0)
C = np.array([[0., -0.23], [0.83, .23]])
X = np.r_[np.dot(np.random.randn(n, dim), C),
np.dot(np.random.randn(n, dim), C) + np.array([1, 1])]
y = np.hstack((np.zeros(n), np.ones(n)))
return X, y
def dataset_cov():
'''Generate 2 Gaussians samples with different covariance matrices'''
n, dim = 300, 2
np.random.seed(0)
C = np.array([[0., -1.], [2.5, .7]]) * 2.
X = np.r_[np.dot(np.random.randn(n, dim), C),
np.dot(np.random.randn(n, dim), C.T) + np.array([1, 4])]
y = np.hstack((np.zeros(n), np.ones(n)))
return X, y
###############################################################################
# plot functions
def plot_data(lda, X, y, y_pred, fig_index):
splot = plt.subplot(2, 2, fig_index)
if fig_index == 1:
plt.title('Linear Discriminant Analysis')
plt.ylabel('Data with fixed covariance')
elif fig_index == 2:
plt.title('Quadratic Discriminant Analysis')
elif fig_index == 3:
plt.ylabel('Data with varying covariances')
tp = (y == y_pred) # True Positive
tp0, tp1 = tp[y == 0], tp[y == 1]
X0, X1 = X[y == 0], X[y == 1]
X0_tp, X0_fp = X0[tp0], X0[~tp0]
X1_tp, X1_fp = X1[tp1], X1[~tp1]
alpha = 0.5
# class 0: dots
plt.plot(X0_tp[:, 0], X0_tp[:, 1], 'o', alpha=alpha,
color='red')
plt.plot(X0_fp[:, 0], X0_fp[:, 1], '*', alpha=alpha,
color='#990000') # dark red
# class 1: dots
plt.plot(X1_tp[:, 0], X1_tp[:, 1], 'o', alpha=alpha,
color='blue')
plt.plot(X1_fp[:, 0], X1_fp[:, 1], '*', alpha=alpha,
color='#000099') # dark blue
# class 0 and 1 : areas
nx, ny = 200, 100
x_min, x_max = plt.xlim()
y_min, y_max = plt.ylim()
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx),
np.linspace(y_min, y_max, ny))
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
plt.pcolormesh(xx, yy, Z, cmap='red_blue_classes',
norm=colors.Normalize(0., 1.))
plt.contour(xx, yy, Z, [0.5], linewidths=2., colors='k')
# means
plt.plot(lda.means_[0][0], lda.means_[0][1],
'o', color='black', markersize=10)
plt.plot(lda.means_[1][0], lda.means_[1][1],
'o', color='black', markersize=10)
return splot
def plot_ellipse(splot, mean, cov, color):
v, w = linalg.eigh(cov)
u = w[0] / linalg.norm(w[0])
angle = np.arctan(u[1] / u[0])
angle = 180 * angle / np.pi # convert to degrees
# filled Gaussian at 2 standard deviation
ell = mpl.patches.Ellipse(mean, 2 * v[0] ** 0.5, 2 * v[1] ** 0.5,
180 + angle, facecolor=color, edgecolor='yellow',
linewidth=2, zorder=2)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.5)
splot.add_artist(ell)
splot.set_xticks(())
splot.set_yticks(())
def plot_lda_cov(lda, splot):
plot_ellipse(splot, lda.means_[0], lda.covariance_, 'red')
plot_ellipse(splot, lda.means_[1], lda.covariance_, 'blue')
def plot_qda_cov(qda, splot):
plot_ellipse(splot, qda.means_[0], qda.covariances_[0], 'red')
plot_ellipse(splot, qda.means_[1], qda.covariances_[1], 'blue')
###############################################################################
for i, (X, y) in enumerate([dataset_fixed_cov(), dataset_cov()]):
# Linear Discriminant Analysis
lda = LinearDiscriminantAnalysis(solver="svd", store_covariance=True)
y_pred = lda.fit(X, y).predict(X)
splot = plot_data(lda, X, y, y_pred, fig_index=2 * i + 1)
plot_lda_cov(lda, splot)
plt.axis('tight')
# Quadratic Discriminant Analysis
qda = QuadraticDiscriminantAnalysis(store_covariances=True)
y_pred = qda.fit(X, y).predict(X)
splot = plot_data(qda, X, y, y_pred, fig_index=2 * i + 2)
plot_qda_cov(qda, splot)
plt.axis('tight')
plt.suptitle('Linear Discriminant Analysis vs Quadratic Discriminant Analysis')
plt.show()
LDA与PCA,LDA与PCA都可以借助SVD求解,但本质是不同的:
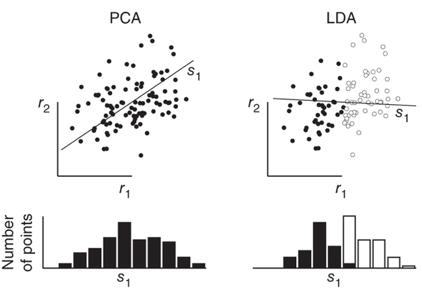
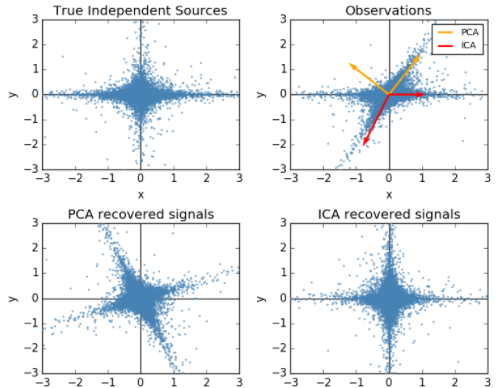
顺便提一句之前梳理的独立成分分析(ICA)与PCA的差别,PCA立足点是相关性,是基于协方差矩阵(二阶统计量);而ICA立足点是独立性,利用概率分布(也就是高阶统计量),当然如果是正态分布,二者就等价了。
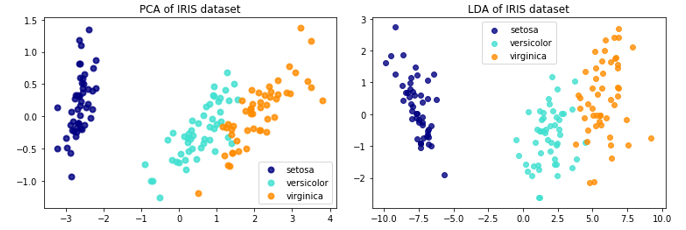
关于LDA、PCA降维的对比,Sklearn给出了IRIS数据的示例:
The Iris dataset represents 3 kind of Iris flowers (Setosa, Versicolour and Virginica) with 4 attributes: sepal length, sepal width, petal length and petal width.即数据类别数是3,每一个样本对应的特征维度是4,现在分别用LDA、PCA降至2维。
code:
print(__doc__)
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.decomposition import PCA
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
iris = datasets.load_iris()
X = iris.data
y = iris.target
target_names = iris.target_names
pca = PCA(n_components=2)
X_r = pca.fit(X).transform(X)
lda = LinearDiscriminantAnalysis(n_components=2)
X_r2 = lda.fit(X, y).transform(X)
# Percentage of variance explained for each components
print('explained variance ratio (first two components): %s'
% str(pca.explained_variance_ratio_))
plt.figure()
colors = ['navy', 'turquoise', 'darkorange']
lw = 2
for color, i, target_name in zip(colors, [0, 1, 2], target_names):
plt.scatter(X_r[y == i, 0], X_r[y == i, 1], color=color, alpha=.8, lw=lw,
label=target_name)
plt.legend(loc='best', shadow=False, scatterpoints=1)
plt.title('PCA of IRIS dataset')
plt.figure()
for color, i, target_name in zip(colors, [0, 1, 2], target_names):
plt.scatter(X_r2[y == i, 0], X_r2[y == i, 1], alpha=.8, color=color,
label=target_name)
plt.legend(loc='best', shadow=False, scatterpoints=1)
plt.title('LDA of IRIS dataset')
plt.show()
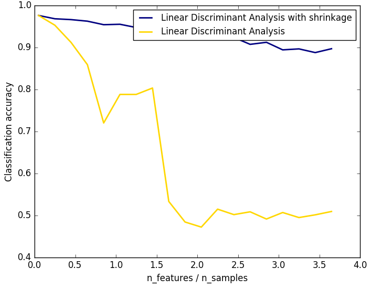
Sklearn还提供了归一化:

只在求解方法为lsqr和eigen时有效,就是前面提到的特征值分解和最小二乘啦。
分别用归一化/不归一化:
clf1 = LinearDiscriminantAnalysis(solver='lsqr', shrinkage='auto').fit(X, y) clf2 = LinearDiscriminantAnalysis(solver='lsqr', shrinkage=None).fit(X, y)
应用实例:
from __future__ import division
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_blobs
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
n_train = 20 # samples for training
n_test = 200 # samples for testing
n_averages = 50 # how often to repeat classification
n_features_max = 75 # maximum number of features
step = 4 # step size for the calculation
def generate_data(n_samples, n_features):
"""Generate random blob-ish data with noisy features.
This returns an array of input data with shape `(n_samples, n_features)`
and an array of `n_samples` target labels.
Only one feature contains discriminative information, the other features
contain only noise.
"""
X, y = make_blobs(n_samples=n_samples, n_features=1, centers=[[-2], [2]])
# add non-discriminative features
if n_features > 1:
X = np.hstack([X, np.random.randn(n_samples, n_features - 1)])
return X, y
acc_clf1, acc_clf2 = [], []
n_features_range = range(1, n_features_max + 1, step)
for n_features in n_features_range:
score_clf1, score_clf2 = 0, 0
for _ in range(n_averages):
X, y = generate_data(n_train, n_features)
clf1 = LinearDiscriminantAnalysis(solver='lsqr', shrinkage='auto').fit(X, y)
clf2 = LinearDiscriminantAnalysis(solver='lsqr', shrinkage=None).fit(X, y)
X, y = generate_data(n_test, n_features)
score_clf1 += clf1.score(X, y)
score_clf2 += clf2.score(X, y)
acc_clf1.append(score_clf1 / n_averages)
acc_clf2.append(score_clf2 / n_averages)
features_samples_ratio = np.array(n_features_range) / n_train
plt.plot(features_samples_ratio, acc_clf1, linewidth=2,
label="Linear Discriminant Analysis with shrinkage", color='navy')
plt.plot(features_samples_ratio, acc_clf2, linewidth=2,
label="Linear Discriminant Analysis", color='gold')
plt.xlabel('n_features / n_samples')
plt.ylabel('Classification accuracy')
plt.legend(loc=1, prop={'size': 12})
plt.suptitle('Linear Discriminant Analysis vs. \
shrinkage Linear Discriminant Analysis (1 discriminative feature)')
plt.show()
结果图可以看出,shrinkage更鲁棒:
参考
- http://blog.csdn.net/daunxx/article/details/51881956
- http://scikit-learn.org/stable/modules/lda_qda.html



