nature method for the first exon alternative splicing
《A paired-end sequencing strategy to map the complex landscape of transcription initiation》
《Tissue-Specific and Ubiquitous Expression Patterns from Alternative Promoters of Human Genes》
第一篇:来自nature method
用的是果蝇的胚胎RNASeq数据分析了其TSS部位的initiation landscape,也就是本人所感兴趣的AFE,在哺乳动物体内也发现了可以用特定motif表征的相似的启动子部位特征。再有,他们发现了5‘帽子端的转录起源于编码的外显子,从本文章中看来,他们的发现TSS的存在并不是形成转录本的过程中选择性剪切的结果,而是转录前的基因coding region的改造作用所产生的产物,证明了paired-end tss analysis to be a powerful method to uncover the transcriptional complexity of eukaryotic genomes.
其中想说的是作者的开篇:
recent studies using high-throughput sequencing protocols have uncovered the complexity of mammalian transcription by rnA polymerase ii, helping to de ne several initiation patterns in which transcription start sites (tsss) cluster in both narrow and broad genomic windows. ![]() 遣词造句还不错
遣词造句还不错
key words:
TSS:Central to this process is the core promoter region of ~100 nucleotides (nt) surrounding the transcription start site (TSS) of a gene
TSS区域的作用是其间DNA motif sequence 保证了RNA转录酶的召集,从而启动转录,
CAGE:the capped analysis of gene expression (CAGE) protocol has been used to generate comprehensive
mammalian libraries of short sequence tags, (CAGE的一般作用)which have led to the identification of distinct transcription initiation patterns
TATA box:an over representation of the canonical TATA box sequence motif in ‘singlepeak’ promoters and CpG islands overlapping ‘broad range’ promoters
目前5‘端的seqing technology有CAGE protocol,deep CAGE protocol,PEAT(本篇作者团队开发的,杜克大学药学院)
PEAT:paired-end analysis of TSSs
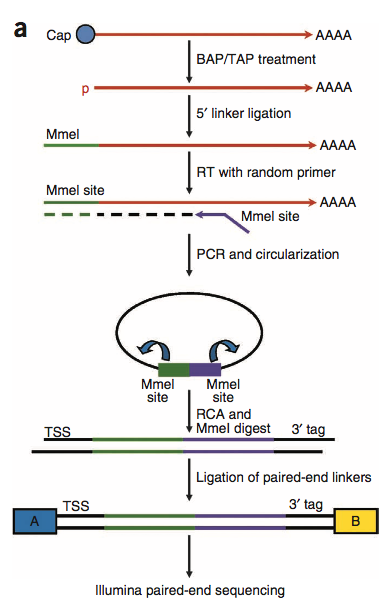
PEAT的strategy图示:RCA:rolling circle amplification(特色)
##########################
3′ reads were mostly mapped to coding regions of annotated genes, indicating the success of the pairedend library construction.
mapping之后的reads类型分布,类型不仅仅显示了3’、5’的测序结果异同,多少也显示了测序深度的大小。3’端数据大多映射到了编码区域也反映了双端建库的成功。
Characterization of read clusters and initiation patterns:TSS clusters and initiation patterns identified in the Drosophila embryo.
a图揭示了5‘端的TSS Tags 组成的density estimate plot,并且TSS 包含95% of the reads的cluster被密集的表示在黑线以及棕黄色area。
b图是所有大于100bpreads的cluster的基因位置饼图,分类的依据则是相应转录组的注释位置以及所给cluster分配的模式。
c图很直观,d图:TSS的窄峰、宽峰、弱峰的分布情况
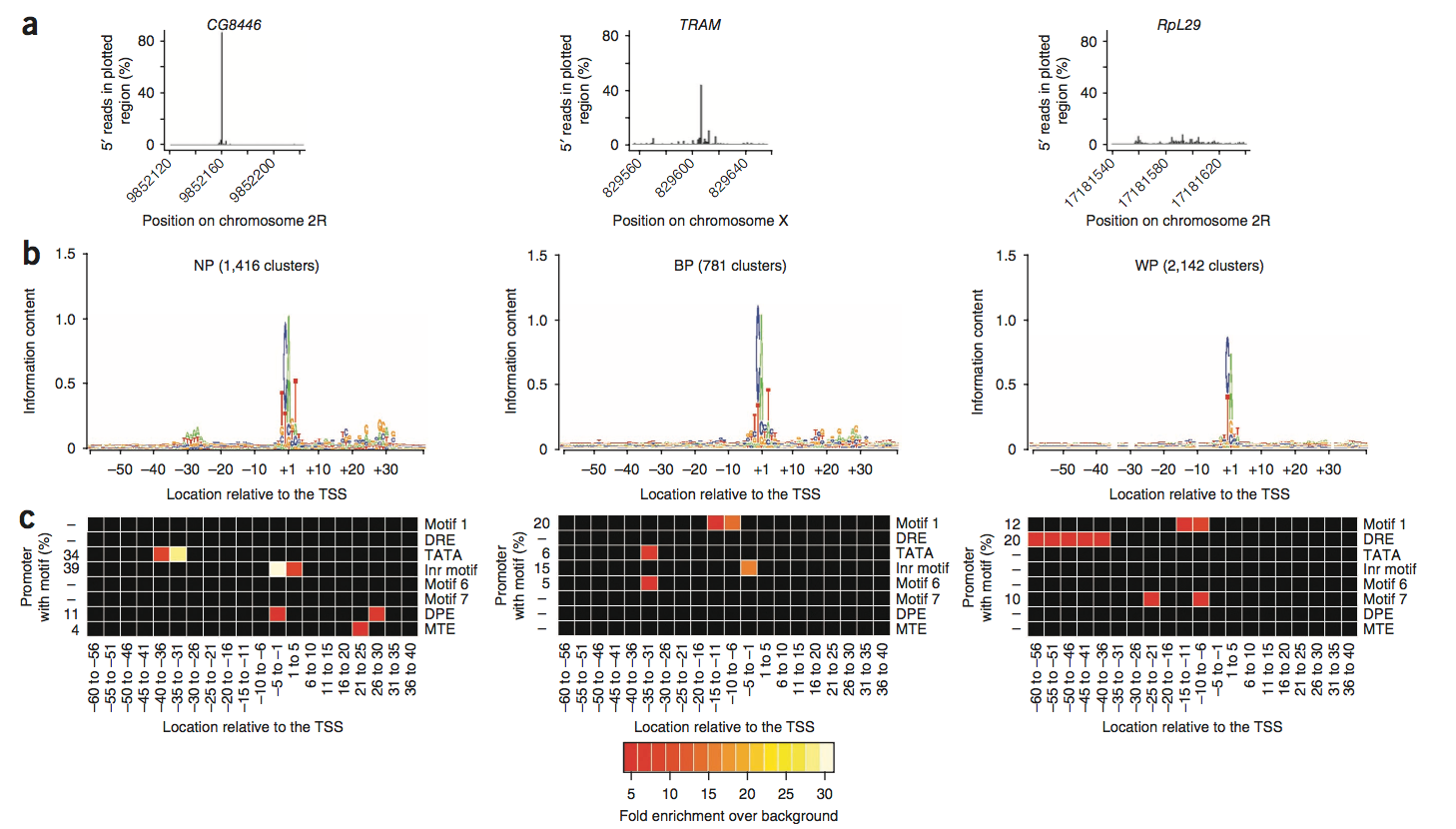
Promoter motifs associated with distinct promoter types:Initiation patterns are linked to specific core promoter
In mammals, ‘single-peak’ and ‘broad-range’ promoters tend to be associated with TATA box sequence and CpG islands As the fly genome does not contain CpG islands, our identification of BP and WP promoters was intriguing
#######
Unlike mammalian WP promoters, which have been associated with CpG islands,Drosophila WP promoters were strongly associated with three motifs (motif 1, DRE and motif 7):果蝇种属中的weak promoter不似哺乳动物中的weak peak与CpG islands呈很高的相关性,而是与三种motifs有很高的相关性。
BP promoters, which have characteristics of both NP and WP promoters, showed a combination of the most frequent motifs found in the other promoter types。而对于broad peak的 promoter的motifs则是结合NP以及WP的motifs的结合体,拥有两种promoters的motifs的co-features
其中:
The Inr motif and motif 1 had in common a strongly conserved ‘TCA’ trinucleotide (the minimal initiator consensus sequence,而TCA又是最小启动子一致序列).
Likewise, motif 6 was enriched at the same location as the TATA box and contains a minimal TAT consensus sequence that is also present in the canonical TATA motif, suggesting that motif 6 and motif 1 combination is an alternative to the classic TATA box and Inr motif pair
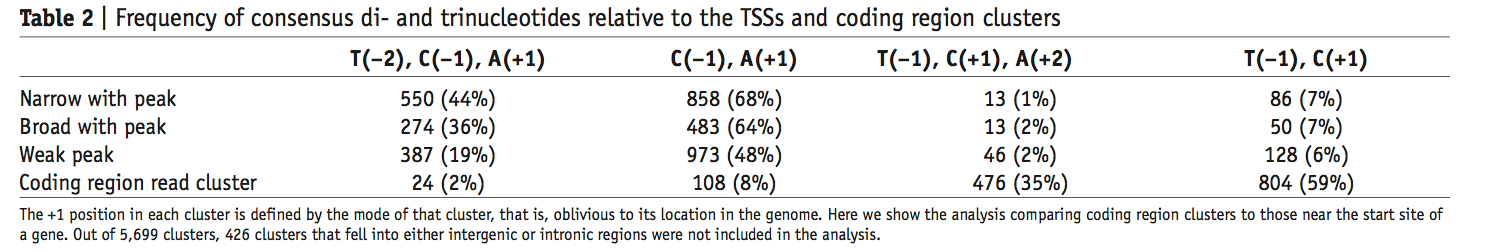
总之,本文总结了几种5‘端的capping转录本的机制:
First, they might result from bona fide start sites in the coding region
Alternatively, these transcripts may be derived from longer precursors, for which the internal cap is introduced posttranscriptionally by a recapping mechanism
our data suggested that 5-capped coding clusters are unlikely initiated by RNA polymerase II
Moreover, the locations of the 5-coding region clusters spread evenly across the exons except for a lack of clusters at the 3-end of the exon
In the PEAT dataset, we observed that the downstream paired tags of the coding clusters were predominantly located in well-annotated exons rather than introns (~100-fold enrichment), indicating that internally capped transcripts were spliced or at least partially spliced(5‘capped 帽子是剪切机制导致的存在而不是转录后的事件).



