some transcripts quantification brief comprehensions


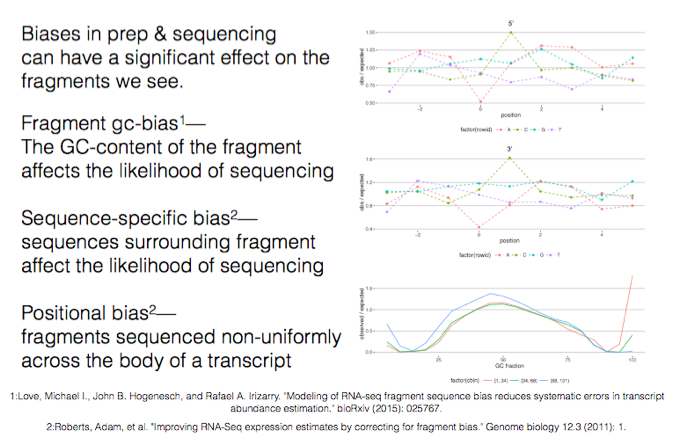
Some biases in the standard rnaseq analysis
有参拼接:Stringtie 、 Cufflinks and Traph
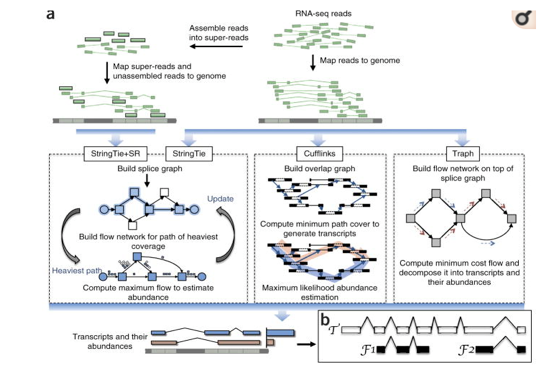
flow network algorithm : maximal and minimal methods respectively
无参组装:Trinity
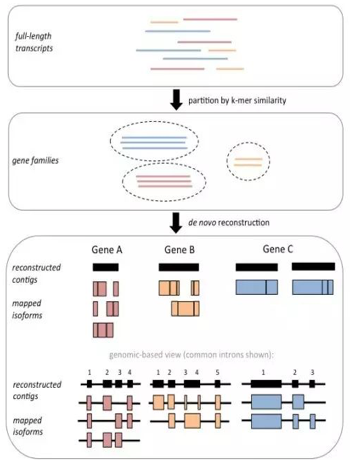
clustering by K-mers
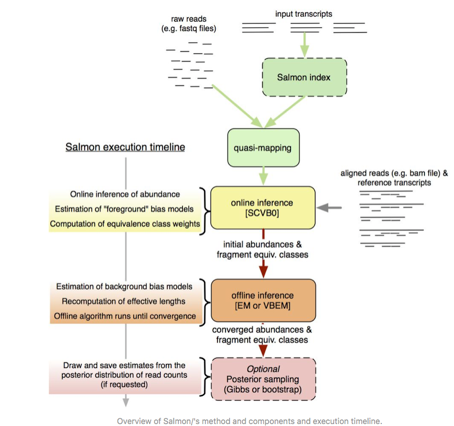
salmon: as for now the best performance software
DAG:有向无环图
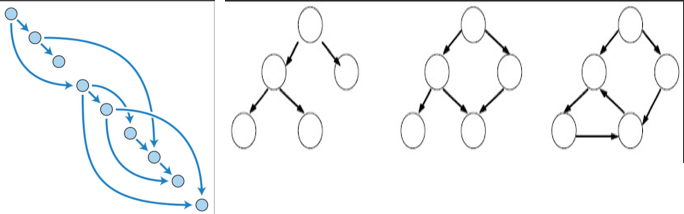

bagging是减少variance,而boosting是减少bias
A- bagging随机选取data的subset,outlier因为比例比较低,参与model training的几率也比较低,所以bagging降低了outliers和noise对model的影响,所以降低了variance。
B-boosting参zh Bright的答案,minimize loss function by definition minimize bias.
Streaming fragment assignment for real-time analysis of sequencing experiments
流形碎片的实时测序实验
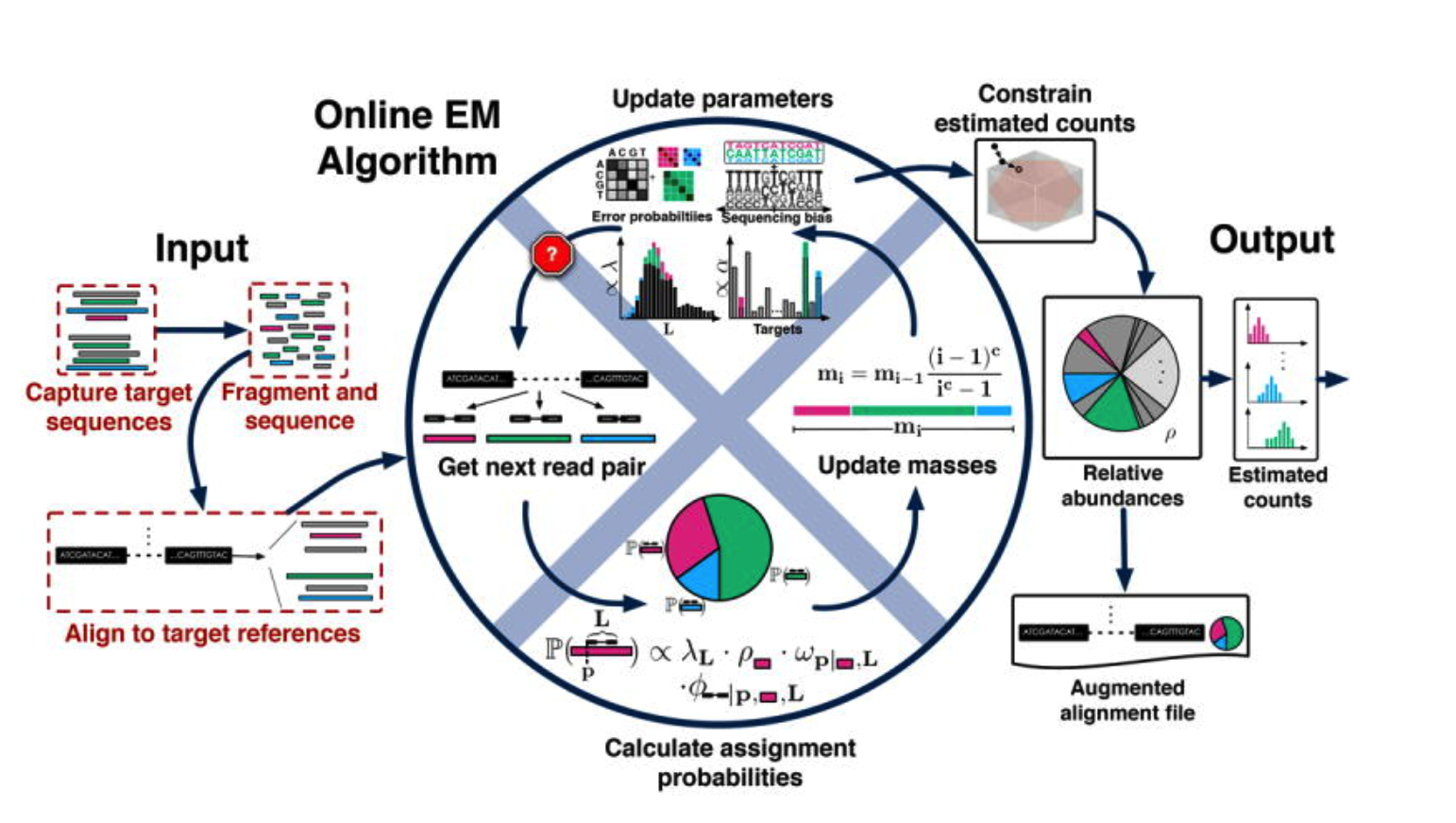
TIPS:
在估计丰度时候很容易用错或者是正确使用与否是很关键的一步:
RPKM:Reads Per Kilobase of exon modelper Million mapped reads (每千个碱基的转录每百万映射读取的reads),主要用来对单端测序(single-end RNA-seq)进行定量的方法。RPKM= total exon reads/ (mapped reads (Millions) * exon length(KB))
FPKM:
Fragments Per Kilobase of exon model per Million mapped fragments(每千个碱基的转录每百万映射读取的fragments),主要是针对pair-end测序表达量进行计算
TPM:
Transcripts Per Kilobase of exonmodel per Million mapped reads (每千个碱基的转录每百万映射读取的Transcripts),优化的RPKM计算方法,可以用于同一物种不同组织的比较。
TPM (推荐软件,RSEM) 的计算公式:
TPMi={( Ni/Li )*1000000 } / sum( Ni/Li+……..+ Nm/Lm )
CPM/RPM:
Reads/Counts of exon model per Million mapped reads (每百万映射读取的reads).
RPM的计算公式:
RPM=total exon reads / mapped reads (Millions)


